5VCZ
 
 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH Bosutinib isomer | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3,5-DICHLORO-4-METHOXYPHENYL)AMINO]-6-METHOXY-7-[3-(4-METHYLPIPERAZIN-1-YL)PROPOXY]QUINOLINE-3-CARBONITRILE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5O59
 
 | | Cellobiohydrolase Cel7A from T. atroviride | | Descriptor: | 1-thio-beta-D-glucopyranose, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Borisova, A.S, Stahlberg, J, Hansson, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Correlation of structure, function and protein dynamics in GH7 cellobiohydrolases from Trichoderma atroviride, T. reesei and T. harzianum.
Biotechnol Biofuels, 11, 2018
|
|
7FZN
 
 | | Crystal Structure of human FABP4 in complex with (1S,2S,5R)-3-(2-thiophen-3-ylacetyl)-3-azabicyclo[3.1.0]hexane-2-carboxylic acid, i.e. SMILES OC(=O)[C@@H]1[C@H]2C[C@H]2CN1C(=O)Cc3ccsc3 with IC50=1.7 microM | | Descriptor: | (1R,2S,5S)-3-[(thiophen-3-yl)acetyl]-3-azabicyclo[3.1.0]hexane-2-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6RS0
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3S,4R,5R)-1-(2-(Benzyloxy)ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol | | Descriptor: | (2~{S},3~{S},4~{R})-2-(hydroxymethyl)-1-(2-phenylmethoxyethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
6M8Q
 
 | | Cleavage and Polyadenylation Specificity Factor Subunit 3 (CPSF3) in complex with NVP-LTM531 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 3, N-{3,5-dichloro-2-hydroxy-4-[2-(4-methylpiperazin-1-yl)ethoxy]benzene-1-carbonyl}-L-phenylalanine, PHOSPHATE ION, ... | | Authors: | Weihofen, W.A, Salcius, M, Michaud, G. | | Deposit date: | 2018-08-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing's sarcoma.
Nat.Chem.Biol., 16, 2020
|
|
7G0Z
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with 5-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]-3,6-dihydro-2H-pyran-4-carboxylic acid, i.e. SMILES S1C(=C(C2=NC(=NO2)C2CC2)C2=C1CCCC2)NC(=O)C1=C(CCOC1)C(=O)O with IC50=0.48537 microM | | Descriptor: | 5-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}-3,6-dihydro-2H-pyran-4-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6TC3
 
 | |
5UXE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P178 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P178
To Be Published
|
|
3AT2
 
 | | Crystal structure of CK2alpha | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha | | Authors: | Kinoshita, T. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A detailed thermodynamic profile of cyclopentyl and isopropyl derivatives binding to CK2 kinase
Mol.Cell.Biochem., 356, 2011
|
|
7FW3
 
 | | Crystal Structure of human FABP4 in complex with 7-(4-chlorophenyl)-1,2,3,4-tetrahydronaphthalene-1-carboxylic acid, i.e. SMILES c12c(ccc(c2)c2ccc(cc2)Cl)CCC[C@H]1C(=O)O with IC50=3.3 microM | | Descriptor: | (1R)-7-(4-chlorophenyl)-1,2,3,4-tetrahydronaphthalene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G1V
 
 | | Crystal Structure of human FABP4 in complex with 3-[(2-sulfanylphenyl)carbamoyl]bicyclo[2.2.1]heptane-2-carboxylic acid, i.e. SMILES [C@@H]1([C@H]2C[C@@H]([C@H]1C(=O)O)CC2)C(=O)Nc1c(cccc1)S with IC50=6.5 microM | | Descriptor: | (1R,2R,3R,4S)-3-(1,3-benzothiazol-2-yl)bicyclo[2.2.1]heptane-2-carboxylic acid, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kumagai, Y, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6HDG
 
 | | D170N variant of beta-phosphoglucomutase from Lactococcus lactis complexed with beta-G1P in a closed conformer to 1.2 A. | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6MD7
 
 | |
7FXA
 
 | | Crystal Structure of human FABP4 in complex with 6-bromo-4-hexyl-2-oxo-1,3-dihydroquinoline-4-carboxylic acid, i.e. SMILES [C@]1(c2c(NC(=O)C1)ccc(c2)Br)(CCCCCC)C(=O)O with IC50=1.2 microM | | Descriptor: | (4S)-6-bromo-4-hexyl-2-oxo-1,2,3,4-tetrahydroquinoline-4-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Hofheinz, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8TKP
 
 | | Structure of the C. elegans TMC-2 complex | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, S, Jeong, H, Goehring, A, Posert, R, Gouaux, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7ULG
 
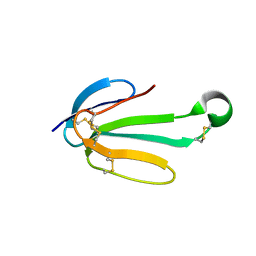 | | recombinant alpha cobra toxin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Alpha-cobratoxin | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2022-04-04 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of recombinant alpha cobra toxin at 1.57 Angstroms
To Be Published
|
|
5V08
 
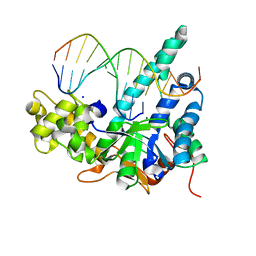 | | Crystal structure of human exonuclease 1 Exo1 (D173A) in complex with 5' recessed-end DNA (rVI) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.812 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6GVG
 
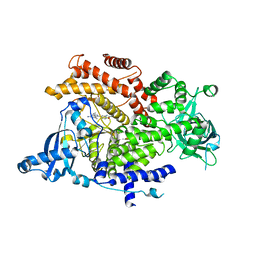 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-4-methyl-1H-pyrazolo[3,4-d]pyrimidin-6-ylamine | | Descriptor: | 5-(6-azanyl-4-methyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7FZY
 
 | | Crystal Structure of human FABP4 in complex with 6-chloro-4-phenyl-2-piperidin-1-yl-3-(1H-tetrazol-5-yl)quinoline, i.e. SMILES c1(ccc2c(c1)c(c(c(n2)N1CCCCC1)C1=NN=NN1)c1ccccc1)Cl with IC50=0.0201184 microM | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-chloro-4-phenyl-2-(piperidin-1-yl)-3-(1H-tetrazol-5-yl)quinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FYT
 
 | | Crystal Structure of human FABP4 in complex with 5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one, i.e. SMILES c1(cccc(c1C)Cl)OCC1=CC(=O)N2N=C(c3ccccc3)N=C2N1 with IC50=0.061 microM | | Descriptor: | (8S)-5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6WLW
 
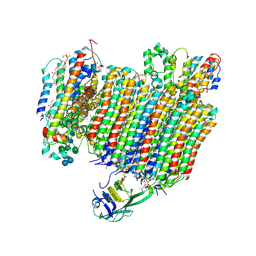 | | The Vo region of human V-ATPase in state 1 (focused refinement) | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, L, Wu, H, Fu, T.-M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
8E73
 
 | | Vigna radiata supercomplex I+III2 (full bridge) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plant-specific features of respiratory supercomplex I + III 2 from Vigna radiata.
Nat.Plants, 9, 2023
|
|
7G1F
 
 | | Crystal Structure of human FABP4 in complex with 1-(4-methylphenyl)sulfonyl-3-(8-tricyclo[5.2.1.02,6]decanyl)urea, i.e. SMILES [C@@H]12[C@H]3[C@@H]([C@H](C2)C[C@H]1NC(=O)NS(=O)(=O)c1ccc(cc1)C)CCC3 with IC50=1.3 microM | | Descriptor: | 4-methyl-N-{[(3aR,4R,5R,7R,7aR)-octahydro-1H-4,7-methanoinden-5-yl]carbamoyl}benzene-1-sulfonamide, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Stempel, A, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7G1L
 
 | | Crystal Structure of human FABP4 in complex with 6-(1,3-benzodioxol-5-ylmethyl)-3-sulfanyl-1,2,4-triazin-5-ol | | Descriptor: | 6-[(2H-1,3-benzodioxol-5-yl)methyl]-3-sulfanyl-1,2,4-triazin-5-ol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5VPO
 
 | | The 70S P-site ASL SufA6 complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hong, S, Sunita, S, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2017-05-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Mechanism of tRNA-mediated +1 ribosomal frameshifting.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
