1PKW
 
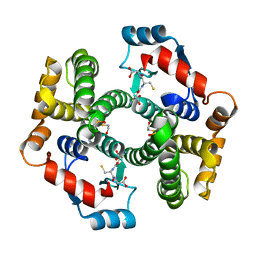 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with glutathione | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PL1
 
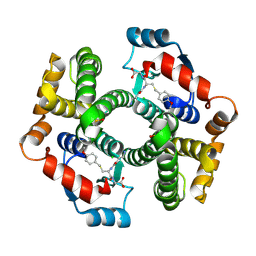 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IX6
 
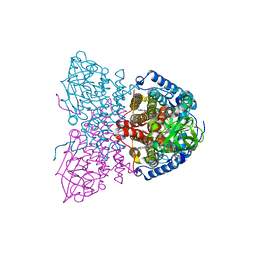 | | SHORT CHAIN SPECIFIC ACYL-COA OXIDASE FROM ARABIDOPSIS THALIANA, ACX4 | | Descriptor: | ACYL-COENZYME A OXIDASE 4, PEROXISOMAL, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mackenzie, J, Pedersen, L, Arent, S, Henriksen, A. | | Deposit date: | 2006-07-07 | | Release date: | 2006-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Controlling Electron Transfer in Acyl-Coa Oxidases and Dehydrogenases: A Structural View.
J.Biol.Chem., 281, 2006
|
|
2IGN
 
 | | Crystal structure of recombinant pyranose 2-oxidase H167A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Divne, C. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate binding and regioselective oxidation of monosaccharides at c3 by pyranose 2-oxidase.
J.Biol.Chem., 281, 2006
|
|
2IGO
 
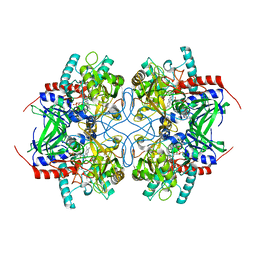 | |
5FL4
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, ACETIC ACID, CARBONIC ANHYDRASE 9, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
2JGI
 
 | |
2JKG
 
 | | Plasmodium falciparum profilin | | Descriptor: | MAGNESIUM ION, OCTAPROLINE PEPTIDE, PROFILIN | | Authors: | Kursula, I, Kursula, P, Ganter, M, Panjikar, S, Matuschewski, K, Schueler, H. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis for Parasite-Specific Functions of the Divergent Profilin of Plasmodium Falciparum.
Structure, 16, 2008
|
|
2JAU
 
 | | Crystal structure of D41N variant of human mitochondrial 5'(3')- deoxyribonucleotidase (mdN) in complex with 3'-azidothymidine 5'- monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, 5'(3')-DEOXYRIBONUCLEOTIDASE, MAGNESIUM ION, ... | | Authors: | Wallden, K, Ruzzenente, B, Bianchi, V, Nordlund, P. | | Deposit date: | 2006-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Human and Murine Deoxyribonucleotidases: Insights Into Recognition of Substrates and Nucleotide Analogues.
Biochemistry, 46, 2007
|
|
2JKF
 
 | | Plasmodium falciparum profilin | | Descriptor: | PROFILIN | | Authors: | Kursula, I, Kursula, P, Ganter, M, Panjikar, S, Matuschewski, K, Schueler, H. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Parasite-Specific Functions of the Divergent Profilin of Plasmodium Falciparum
Structure, 16, 2008
|
|
4R3C
 
 | | Crystal structure of p38 alpha MAP kinase in complex with a novel isoform selective drug candidate | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 6-(4-methylpiperazin-1-yl)-3-(naphthalen-2-yl)-4-(pyridin-4-yl)pyridazine, CHLORIDE ION, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting human central nervous system protein kinases: An isoform selective p38 alpha MAPK inhibitor that attenuates disease progression in Alzheimer's disease mouse models.
ACS Chem Neurosci, 6, 2015
|
|
1PL2
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 T68E mutant in complex with decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2JGF
 
 | | Crystal structure of mouse acetylcholinesterase inhibited by non-aged fenamiphos | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL, ... | | Authors: | Hornberg, A, Tunemalm, A.-K, Ekstrom, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Acetylcholinesterase in Complex with Organophosphorus Compounds Suggest that the Acyl Pocket Modulates the Aging Reaction by Precluding the Formation of the Trigonal Bipyramidal Transition State.
Biochemistry, 46, 2007
|
|
5FL6
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-(4-Methylphenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 5-[1-(4-methylphenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, ACETIC ACID, CARBONIC ANHYDRASE IX, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
2JGK
 
 | | Crystal structure of mouse acetylcholinesterase inhibited by aged fenamiphos | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Hornberg, A, Tunemalm, A.-K, Ekstrom, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Acetylcholinesterase in Complex with Organophosphorus Compounds Suggest that the Acyl Pocket Modulates the Aging Reaction by Precluding the Formation of the Trigonal Bipyramidal Transition State.
Biochemistry, 46, 2007
|
|
1PKZ
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PUO
 
 | | Crystal structure of Fel d 1- the major cat allergen | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Major allergen I polypeptide, fused chain 2, ... | | Authors: | Kaiser, L, Gronlund, H, Sandalova, T, Ljunggren, H.G, van Hage-Hamsten, M, Achour, A, Schneider, G. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the major cat allergen Fel d 1, a member of the secretoglobin family.
J.Biol.Chem., 278, 2003
|
|
1S7W
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
5FL5
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-(4-Methoxyphenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[1-(4-methoxyphenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, CARBONIC ANHYDRASE IX, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
2JF0
 
 | | Mus musculus acetylcholinesterase in complex with tabun and Ortho-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
2JGE
 
 | |
2JEY
 
 | | Mus musculus acetylcholinesterase in complex with HLo-7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
2JEZ
 
 | | Mus musculus acetylcholinesterase in complex with tabun and HLo-7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
1TDQ
 
 | | Structural basis for the interactions between tenascins and the C-type lectin domains from lecticans: evidence for a cross-linking role for tenascins | | Descriptor: | Aggrecan core protein, CALCIUM ION, Tenascin-R | | Authors: | Lundell, A, Olin, A.I, Moergelin, M, al-Karadaghi, S, Aspberg, A, Logan, D.T. | | Deposit date: | 2004-05-24 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for interactions between tenascins and lectican C-type lectin domains: evidence for a crosslinking role for tenascins
Structure, 12, 2004
|
|
1T4C
 
 | | Formyl-CoA Transferase in complex with Oxalyl-CoA | | Descriptor: | COENZYME A, Formyl-CoA:oxalate CoA-transferase, OXALIC ACID | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
