3LP9
 
 | | Crystal structure of LS24, A Seed Albumin from Lathyrus sativus | | Descriptor: | CALCIUM ION, CHLORIDE ION, LS-24, ... | | Authors: | Gaur, V, Qureshi, I.A, Singh, A, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional insights of hemopexin fold protein from grass pea
Plant Physiol., 152, 2010
|
|
9DRV
 
 | | Crystal structure of M. tuberculosis PheRS-tRNA complex bound to inhibitor D-004 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Gade, P, Chang, C, Forte, B, Wower, J, Gilbert, I.H, Baragana, B, Michalska, K, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-26 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
3IBH
 
 | | Crystal structure of Saccharomyces cerevisiae Gtt2 in complex with glutathione | | Descriptor: | GLUTATHIONE, Saccharomyces cerevisiae Gtt2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Bao, R, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2009-07-15 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
1WLU
 
 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, phenylacetic acid degradation protein PaaI | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
2XEY
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 3-(1H-INDOL-3-YL)-6-(1H-PYRAZOL-4-YL)IMIDAZO[1,2-A]PYRAZINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, McHardy, T, Klair, S, Boxall, K, Fisher, M, Cherry, M, Allen, C.E, Addison, G.J, Ellard, J, Aherne, G.W, Westwood, I.M, van Montfort, R, Garrett, M.D, Reader, J.C, Collins, I. | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and Evaluation of 3,6-Di(Hetero)Aryl Imidazo[1,2-A]Pyrazines as Inhibitors of Checkpoint and Other Kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7AQ1
 
 | | Crystal structure of human mature meprin beta in complex with the specific inhibitor MWT-S-270 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Linnert, M, Parthier, C, Fritz, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure and Dynamics of Meprin beta in Complex with a Hydroxamate-Based Inhibitor.
Int J Mol Sci, 22, 2021
|
|
1MN0
 
 | | Crystal structure of galactose mutarotase from lactococcus lactis complexed with D-xylose | | Descriptor: | Aldose 1-epimerase, NICKEL (II) ION, alpha-D-xylopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
4ACT
 
 | | CRYSTAL STRUCTURE OF TRANSTHYRETIN IN COMPLEX WITH LIGAND C-17 | | Descriptor: | 3-hydroxy-4-phenoxybenzaldehyde, TRANSTHYRETIN | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Inhibitors: Unique Mechanism of Negative-Cooperativity between Two T4 Binding Sites
To be Published
|
|
2XF0
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 3-PHENYL-6-(1H-PYRAZOL-4-YL)IMIDAZO[1,2-A]PYRAZINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, McHardy, T, Klair, S, Boxall, K, Fisher, M, Cherry, M, Allen, C.E, Addison, G.J, Ellard, J, Aherne, G.W, Westwood, I.M, van Montfort, R, Garrett, M.D, Reader, J.C, Collins, I. | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of 3,6-Di(Hetero)Aryl Imidazo[1,2-A]Pyrazines as Inhibitors of Checkpoint and Other Kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6NX9
 
 | |
1WBQ
 
 | | ZnMg substituted aminopeptidase P from E. coli | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, XAA-PRO AMINOPEPTIDASE, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-11-05 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
6A8B
 
 | | Ribokinase from Leishmania donovani with AMPPCP | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Gatreddi, S, Pillalamarri, V, Qureshi, I.A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
2BTP
 
 | | 14-3-3 Protein Theta (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN TAU, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Elkins, J.M, Johansson, A.C.E, Smee, C, Yang, X, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-05 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1N7D
 
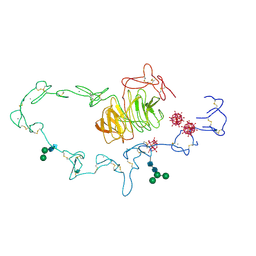 | | Extracellular domain of the LDL receptor | | Descriptor: | 12-TUNGSTOPHOSPHATE, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Rudenko, G, Henry, L, Henderson, K, Ichtchenko, K, Brown, M.S, Goldstein, J.L, Deisenhofer, J. | | Deposit date: | 2002-11-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the LDL receptor extracellular domain at endosomal pH
Science, 298, 2002
|
|
3HLD
 
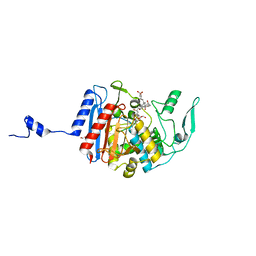 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant complex with monacolin J acid | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, FORMIC ACID, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
6A35
 
 | |
6A34
 
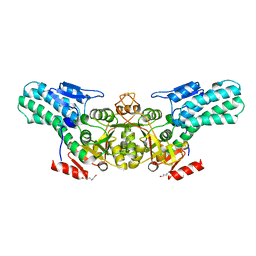 | |
8FA3
 
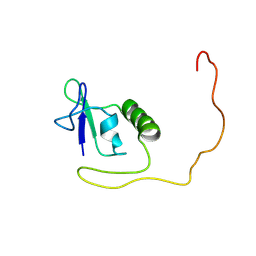 | |
1LBW
 
 | | Crystal Structure of apo-form (P32) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1Q66
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-6-AMINOMETHYL-8-phenylsulfanylmethyl-3H-QUINAZOLIN-4-ONE crystallized at pH 5.5 | | Descriptor: | 2-AMINO-6-AMINOMETHYL-8-PHENYLSULFANYLMETHYL-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
6C0Y
 
 | |
8H53
 
 | | Human asparaginyl-tRNA synthetase in complex with asparagine-AMP | | Descriptor: | 4-AMINO-1,4-DIOXOBUTAN-2-AMINIUM ADENOSINE-5'-MONOPHOSPHATE, Asparagine--tRNA ligase, cytoplasmic, ... | | Authors: | Park, J.S, Han, B.W. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Reaction hijacking inhibition of Plasmodium falciparum asparagine tRNA synthetase.
Nat Commun, 15, 2024
|
|
2FLJ
 
 | | Fatty acid binding protein from locust flight muscle in complex with oleate | | Descriptor: | Fatty acid-binding protein, OLEIC ACID | | Authors: | Luecke, C, Qiao, Y, van Moerkerk, H.T.B, Veerkamp, J.H, Hamilton, J.A. | | Deposit date: | 2006-01-06 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Fatty-acid-binding protein from the flight muscle of Locusta migratoria: evolutionary variations in fatty acid binding.
Biochemistry, 45, 2006
|
|
5UYU
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline-3-azaxanthene compound 12 | | Descriptor: | (5S)-3-(3,6-dihydro-2H-pyran-4-yl)-7-[5-(prop-1-yn-1-yl)pyridin-3-yl]-5'H-spiro[1-benzopyrano[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M, Sickmier, E.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of 2-aminooxazoline 3-azaxanthene beta-amyloid cleaving enzyme (BACE) inhibitors with improved selectivity against Cathepsin D.
Medchemcomm, 8, 2017
|
|
1L5Z
 
 | | CRYSTAL STRUCTURE OF THE E121K SUBSTITUTION OF THE RECEIVER DOMAIN OF SINORHIZOBIUM MELILOTI DCTD | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN DCTD, GLYCEROL, SULFATE ION | | Authors: | Park, S, Meyer, M, Jones, A.D, Yennawar, H.P, Yennawar, N.H, Nixon, B.T. | | Deposit date: | 2002-03-08 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain
FASEB J., 16, 2002
|
|
