7BGG
 
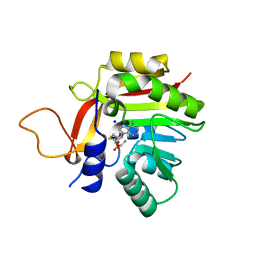 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, heterocyclic toxin methyltransferase (Rv0560c) | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-01-07 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
5AAF
 
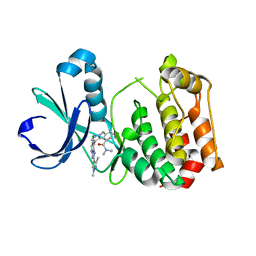 | | Aurora A kinase bound to an imidazopyridine inhibitor (14a) | | Descriptor: | 3-((4-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)-1H-pyrazol-1-yl)methyl)-N,N-dimethylbenzamide, AURORA KINASE A | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7BMK
 
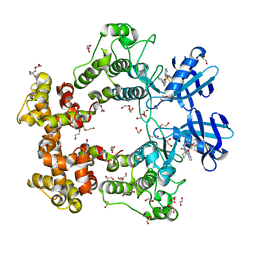 | | ATP-Competitive Partial Antagonists-'PAIR's-Rheostatically Modulate IRE1alpha's Kinase Helix-alphaC to Segregate its RNase-Mediated Biological Outputs | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-tris(fluoranyl)-~{N}-[4-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxynaphthalen-1-yl]ethanesulfonamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feldman, H.C, Ghosh, R, Auyeung, V, Mueller, J.L, Vidadala, V.N, Olivier, A, Backes, B.J, Zikherman, J, Papa, F.R, Maly, D.J. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | ATP-competitive partial antagonists of the IRE1 alpha RNase segregate outputs of the UPR.
Nat.Chem.Biol., 17, 2021
|
|
6I5I
 
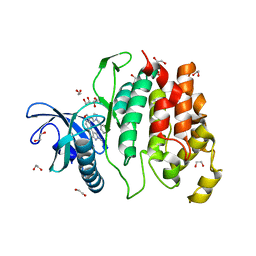 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7XTW
 
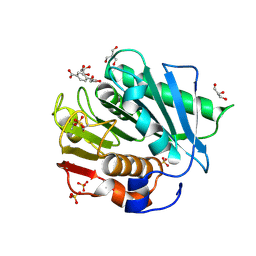 | | The structure of IsPETase in complex with MHET | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
2VZG
 
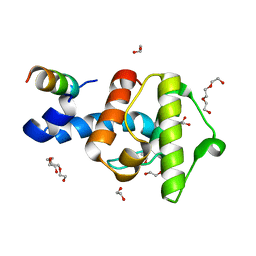 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD2 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
9I7K
 
 | |
4ZX1
 
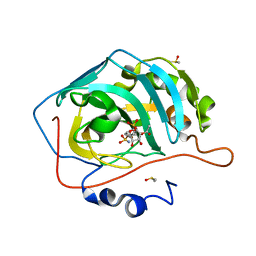 | | Engineered Carbonic Anhydrase IX mimic in complex with a glucosyl sulfamate inhibitor | | Descriptor: | (6R)-5-O-acetyl-2,6-anhydro-6-{[4-(sulfamoyloxy)piperidin-1-yl]sulfonyl}-L-glucitol, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Mahon, B.P, Lomelino, C.L, Salguero, A.L, McKenna, R. | | Deposit date: | 2015-05-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX using Structure-Activity Relationships of Glucosyl-Based Sulfamates
J. Med. Chem., 58, 2015
|
|
6F08
 
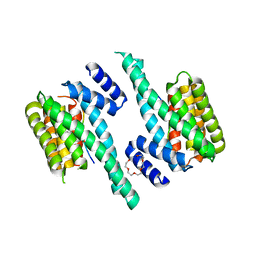 | | 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1) | | Descriptor: | 14-3-3 protein zeta/delta, PENTAETHYLENE GLYCOL, Son of sevenless homolog 1 | | Authors: | Ballone, A, Centorrino, F, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of 14-3-3 zeta in complex with the human Son of sevenless homolog 1 (SOS1).
J. Struct. Biol., 202, 2018
|
|
9BFF
 
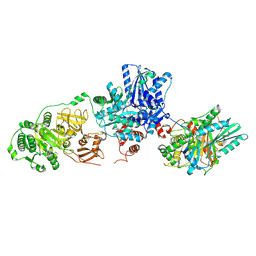 | | Tyrocidine synthetase modules 1 and 2 crosslinked in the condensation state, complex C | | Descriptor: | (7S,10aR)-1-methyl-4-{4-[(5R)-1,1,5-trihydroxy-4,4-dimethyl-1,6,10,15-tetraoxo-2-oxa-7,11,14-triaza-1lambda~5~-phosphahexadecan-16-yl]phenyl}-3,5,6,7,8,9,10,10a-octahydrocycloocta[d]pyridazin-7-yl [(5R)-1,1,5-trihydroxy-4,4-dimethyl-1,6,10-trioxo-2-oxa-7,11-diaza-1lambda~5~-phosphatridecan-13-yl]carbamate (non-preferred name), Tyrocidine synthase 1, Tyrocidine synthase 2 | | Authors: | Heberlig, G.W, Burkart, M.D. | | Deposit date: | 2024-04-17 | | Release date: | 2024-10-09 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Crosslinking intermodular condensation in non-ribosomal peptide biosynthesis.
Nature, 638, 2025
|
|
9IK8
 
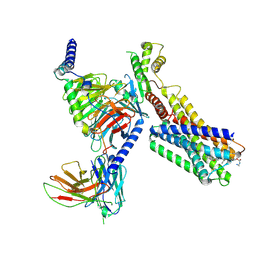 | | Cryo-EM Structure of SSTR1-Gi SST analogs complex | | Descriptor: | DTR-LYS-TY5-PHA-A1D5E-004, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Du, Y. | | Deposit date: | 2024-06-26 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural insights into the binding modes of lanreotide and pasireotide with somatostatin receptor 1.
Acta Pharm Sin B, 15, 2025
|
|
7UT0
 
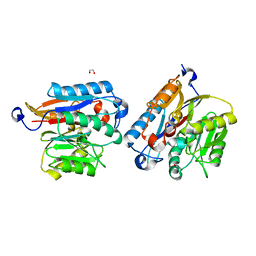 | | Human DDAH-1, apo form | | Descriptor: | 1,2-ETHANEDIOL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Smith, C.A, Ghebre, Y.T. | | Deposit date: | 2022-04-26 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Esomeprazole covalently interacts with the cardiovascular enzyme dimethylarginine dimethylaminohydrolase: Insights into the cardiovascular risk of proton pump inhibitors.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
9IK9
 
 | | Cryo-EM Structure of SST analogs bond SSTR1-Gi complex | | Descriptor: | (4J2)(DCY)(DTY)(DTR)K(DVA)(DCY)(ALO)(NH2), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Du, Y. | | Deposit date: | 2024-06-26 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into the binding modes of lanreotide and pasireotide with somatostatin receptor 1.
Acta Pharm Sin B, 15, 2025
|
|
7S26
 
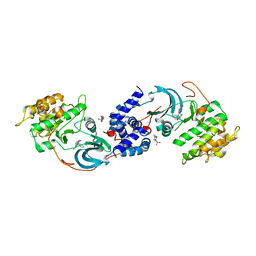 | | ROCK1 IN COMPLEX WITH LIGAND G5018 | | Descriptor: | 2-[methyl(phenyl)amino]-1-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-3,6-dihydropyridin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
9ENP
 
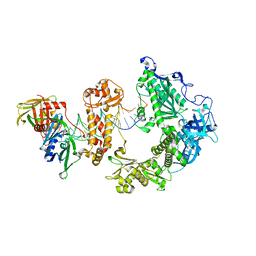 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
6AYK
 
 | |
5KY4
 
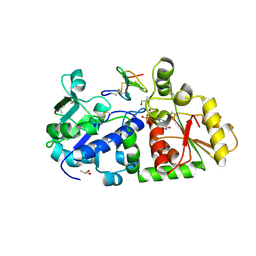 | | mouse POFUT1 in complex with mouse Notch1 EGF26 and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
7BDH
 
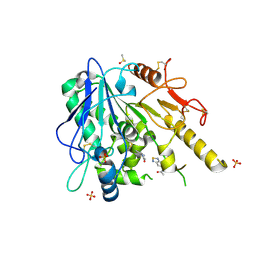 | | Notum Fragment 955 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, N-(2,1,3-Benzothiadiazol-5-yl)acetamide, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BDB
 
 | | Notum Fragment 916 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-ethyl-5-methyl-N-(5-methyl-1,2-oxazol-3-yl)-1,2-oxazole-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BFA
 
 | |
7B98
 
 | | Notum Fragment 282 | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B9U
 
 | | Notum Fragment 609 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(furan-2-yl)methyl]-1lambda~6~,4-thiazinane-1,1-dione, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCF
 
 | | Notum Fragment 722 | | Descriptor: | 2-[4-(2,5-Dioxopyrrolidin-1-yl)phenoxy]acetate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
5AAE
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14d) | | Descriptor: | 3-((4-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)-1H-pyrazol-1-yl)methyl)-5-methylisoxazole, AURORA KINASE A | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7BCC
 
 | | Notum Fragment 705 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(4-pyrrol-1-ylphenyl)morpholine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
