3B0V
 
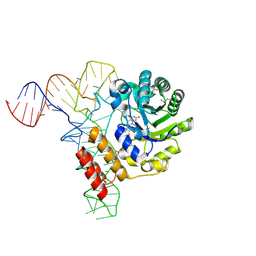 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1U2A
 
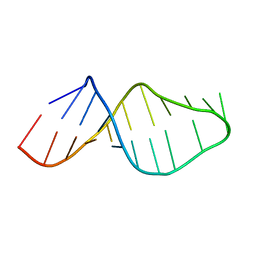 | |
3K4E
 
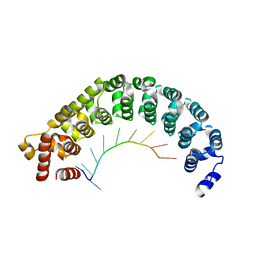 | | Puf3 RNA binding domain bound to Cox17 RNA 3' UTR recognition sequence site A | | Descriptor: | RNA (5'-R(P*CP*UP*UP*GP*UP*AP*UP*AP*UP*A)-3'), mRNA-binding protein PUF3 | | Authors: | Zhu, D, Stumpf, C.R, Krahn, J.M, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2Z67
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) selenium transferase (SepSecS) | | Descriptor: | O-phosphoseryl-tRNA(Sec) selenium transferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Araiso, Y, Ishitani, R, Pailouer, S, Oshikane, H, Domae, N, Soll, D, Nureki, O. | | Deposit date: | 2007-07-23 | | Release date: | 2008-04-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation.
Nucleic Acids Res., 36, 2008
|
|
7E8K
 
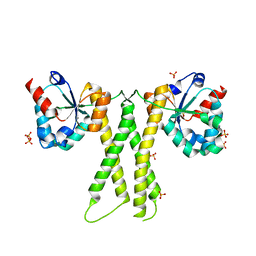 | |
7E8J
 
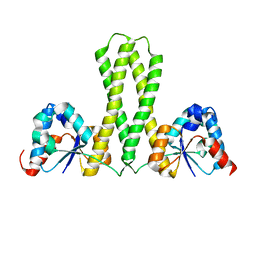 | |
7JL2
 
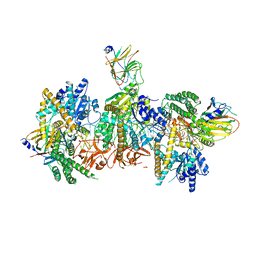 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL0
 
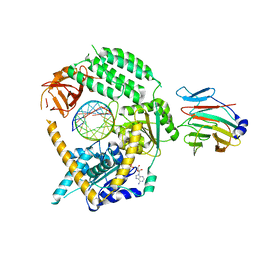 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
5XJ2
 
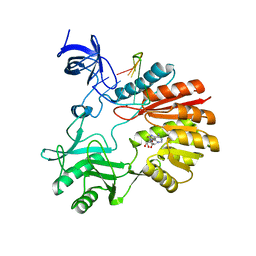 | | Structure of spRlmCD with U747 RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*CP*GP*UP*GP*CP*U)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029, ... | | Authors: | Jiang, Y, Gong, Q. | | Deposit date: | 2017-04-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural insights into substrate selectivity of ribosomal RNA methyltransferase RlmCD
PLoS ONE, 12, 2017
|
|
2KEZ
 
 | | NMR structure of U6 ISL at pH 8.0 | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | Authors: | Venditti, V, Butcher, S.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
2MF0
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer L of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF1
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
4V34
 
 | | The Structure of A-PGS from Pseudomonas aeruginosa (SeMet derivative) | | Descriptor: | ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CHLORIDE ION, SULFATE ION | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5XAQ
 
 | |
7CBG
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-N-[(E)-4-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]but-2-enyl]-2-(methylamino)-3-oxidanyl-butanamide, Threonine--tRNA ligase, ZINC ION | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
7CBI
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl (2~{S},3~{R})-2-azanyl-3-oxidanyl-butanoate, GLYCEROL, ... | | Authors: | Guo, J, Chen, B, Zhou, H. | | Deposit date: | 2020-06-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
2KF0
 
 | | NMR structure of U6 ISL at pH 7.0 | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*CP*CP*UP*GP*CP*AP*UP*AP*AP*GP*GP*AP*UP*GP*AP*AP*CP*C)-3') | | Authors: | Venditti, V, Butcher, S.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minimum-energy path for a u6 RNA conformational change involving protonation, base-pair rearrangement and base flipping.
J.Mol.Biol., 391, 2009
|
|
9BXE
 
 | |
7D58
 
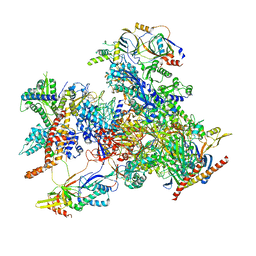 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5GMC
 
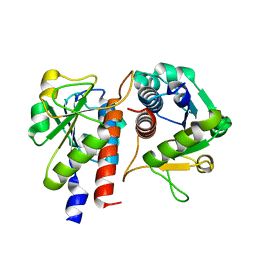 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
5GMB
 
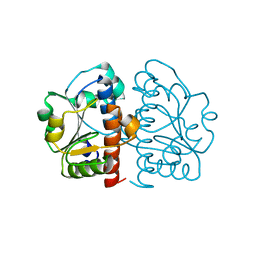 | | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruiginosa | | Descriptor: | tRNA (cytidine/uridine-2'-O-)-methyltransferase TrmJ | | Authors: | Jaroensuk, J, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Liew, C.W, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa.
Nucleic Acids Res., 44, 2016
|
|
9ISV
 
 | |
2K0A
 
 | | 1H, 15N and 13C chemical shift assignments for Rds3 protein | | Descriptor: | Pre-mRNA-splicing factor RDS3, ZINC ION | | Authors: | Loening, N, van Roon, A, Yang, J, Nagai, K, Neuhaus, D. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U2 snRNP protein Rds3p reveals a knotted zinc-finger motif.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3OTC
 
 | |
6KOR
 
 | | Crystal structure of the RRM domain of SYNCRIP | | Descriptor: | Heterogeneous nuclear ribonucleoprotein Q | | Authors: | Chen, Y, Chan, J, Chen, W, Jobichen, C. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | SYNCRIP, a new player in pri-let-7a processing.
Rna, 26, 2020
|
|
