2WD0
 
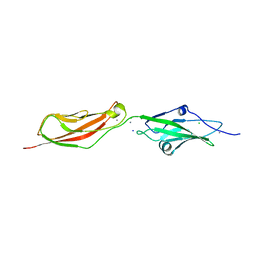 | | CRYSTAL STRUCTURE OF NONSYNDROMIC DEAFNESS (DFNB12) ASSOCIATED MUTANT D124G OF MOUSE CADHERIN-23 EC1-2 | | Descriptor: | CADHERIN-23, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2009-03-18 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Determinants of Cadherin-23 Function in Hearing and Deafness.
Neuron, 66, 2010
|
|
1AKE
 
 | |
3T3I
 
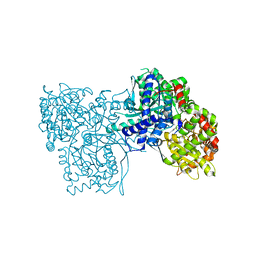 | | Glycogen Phosphorylase b in complex with GlcCF3U | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
148D
 
 | |
3STH
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Toxoplasma gondii | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Staker, B.L, Edwards, T.E, Sankaran, B. | | Deposit date: | 2011-07-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Membrane skeletal association and post-translational allosteric regulation of Toxoplasma gondii GAPDH1.
Mol.Microbiol., 103, 2017
|
|
2VSE
 
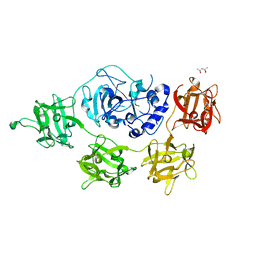 | | Structure and mode of action of a mosquitocidal holotoxin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MOSQUITOCIDAL TOXIN | | Authors: | Treiber, N, Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mode of Action of a Mosquitocidal Holotoxin.
J.Mol.Biol., 381, 2008
|
|
6HA4
 
 | | Crystal structure of PAF - p-sulfonatocalix[4]arene complex | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, GLYCEROL, Pc24g00380 protein | | Authors: | Alex, J.M, Rennie, M, Engilberge, S, Batta, G, Crowley, P.B. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Calixarene-mediated assembly of a small antifungal protein.
Iucrj, 6, 2019
|
|
2FKY
 
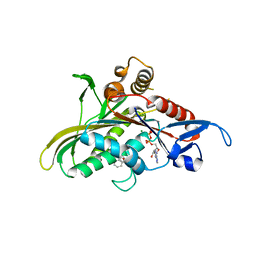 | | crystal structure of KSP in complex with inhibitor 13 | | Descriptor: | (2S)-4-(2,5-DIFLUOROPHENYL)-N-METHYL-2-PHENYL-N-PIPERIDIN-4-YL-2,5-DIHYDRO-1H-PYRROLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 2: the design, synthesis, and characterization of 2,4-diaryl-2,5-dihydropyrrole inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5DPJ
 
 | | sfGFP double mutant - 133/149 p-ethynyl-L-phenylalanine | | Descriptor: | Green fluorescent protein | | Authors: | Dippel, A.B, Olenginski, G.M, Maurici, N, Liskov, M.T, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2015-09-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the effectiveness of spectroscopic reporter unnatural amino acids: a structural study.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2VSK
 
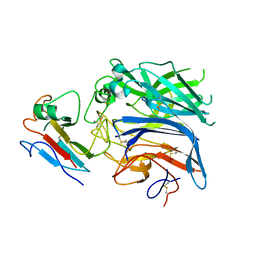 | | Hendra virus attachment glycoprotein in complex with human cell surface receptor ephrinB2 | | Descriptor: | EPHRIN-B2, HEMAGGLUTININ-NEURAMINIDASE | | Authors: | Bowden, T.A, Aricescu, A.R, Gilbert, R.J, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Nipah and Hendra Virus Attachment to Their Cell-Surface Receptor Ephrin-B2
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VTZ
 
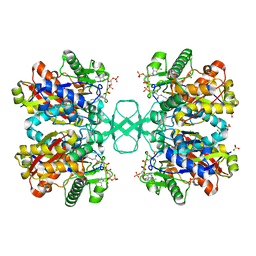 | | Biosynthetic thiolase from Z. ramigera. Complex of the C89A mutant with coenzyme A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
156D
 
 | |
5UHT
 
 | | Structure of the Thermotoga maritima HK853-BeF3-RR468 complex at pH 5.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, Y, Rose, J, Jiang, L, Zhou, P. | | Deposit date: | 2017-01-12 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
1ANP
 
 | |
199L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
3VYV
 
 | | Crystal structure of subtilisin NAT at 1.36 | | Descriptor: | CALCIUM ION, GLYCEROL, Subtilisin NAT | | Authors: | Ushijima, H, Fuchita, N, Kajiwara, T, Motoshima, H, Ueno, G, Watanabe, K. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of subtilisin NAT at 1.36
TO BE PUBLISHED
|
|
1A1Q
 
 | | HEPATITIS C VIRUS NS3 PROTEINASE | | Descriptor: | NS3 PROTEINASE, ZINC ION | | Authors: | Love, R.A, Parge, H.E, Wickersham, J.A, Hostomsky, Z, Habuka, N, Moomaw, E.W, Adachi, T, Hostomska, Z. | | Deposit date: | 1997-12-12 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Cell(Cambridge,Mass.), 87, 1996
|
|
1AB6
 
 | | STRUCTURE OF CHEY MUTANT F14N, V86T | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serranno, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AC1
 
 | | DSBA MUTANT H32L | | Descriptor: | DSBA | | Authors: | Guddat, L.W, Martin, J.L. | | Deposit date: | 1997-02-10 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability.
Protein Sci., 6, 1997
|
|
2WOR
 
 | | co-structure of S100A7 with 1,8 ANS | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, CALCIUM ION, PROTEIN S100-A7, ... | | Authors: | Leon, R, Hof, F, Boulanger, M.J. | | Deposit date: | 2009-07-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Characterization of Binding Sites on S100A7, a Participant in Cancer and Inflammation Pathways.
Biochemistry, 48, 2009
|
|
5TRS
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
3PXT
 
 | | Crystal Structure of Ferrous CO Adduct of MauG in Complex with Pre-Methylamine Dehydrogenase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, CALCIUM ION, ... | | Authors: | Yukl, E.T, Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of CO and NO Adducts of MauG in Complex with Pre-Methylamine Dehydrogenase: Implications for the Mechanism of Dioxygen Activation.
Biochemistry, 50, 2011
|
|
2BF2
 
 | | Crystal structure of native toluene-4-monooxygenase catalytic effector protein, T4moD | | Descriptor: | TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
6HZ4
 
 | | Structure of McrBC without DNA binding domains (one half of the full complex) | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Itoh, Y, Nirwan, N, Saikrishnan, K, Amunts, A. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC.
Nat Commun, 10, 2019
|
|
2X07
 
 | | cytochrome c peroxidase: engineered ascorbate binding site | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murphy, E.J, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-12-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering the substrate specificity and reactivity of a heme protein: creation of an ascorbate binding site in cytochrome c peroxidase.
Biochemistry, 47, 2008
|
|
