7AZ2
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1014 | | Descriptor: | 1-[4-methyl-2-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AOG
 
 | | 14-3-3 sigma in complex with Pin1 binding site pS72 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7AZ1
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1013 | | Descriptor: | 1-[4-methyl-3-(trifluoromethyl)phenyl]-2-phenyl-imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Ottmann, C. | | Deposit date: | 2020-11-14 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
4DAT
 
 | |
7B15
 
 | |
7B13
 
 | |
4DHP
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4E2E
 
 | |
4DHQ
 
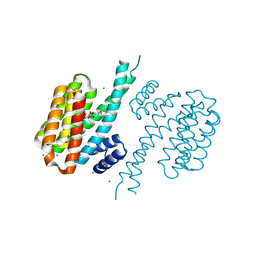 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3-chlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHM
 
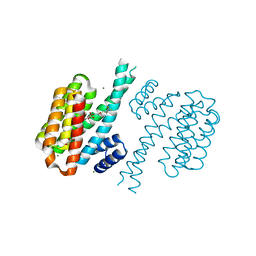 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHT
 
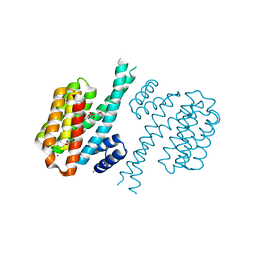 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 PROTEIN SIGMA, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DAU
 
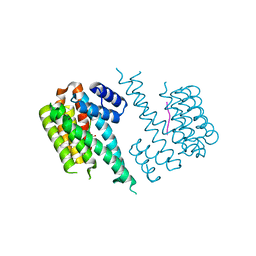 | |
4DX0
 
 | | Structure of the 14-3-3/PMA2 complex stabilized by a pyrazole derivative | | Descriptor: | 14-3-3-like protein E, 4-[(4R)-4-(4-nitrophenyl)-6-oxidanylidene-3-phenyl-1,4-dihydropyrrolo[3,4-c]pyrazol-5-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Richter, A, Rose, R, Hedberg, C, Waldmann, H, Ottmann, C. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Optimised Small-Molecule Stabiliser of the 14-3-3-PMA2 Protein-Protein Interaction.
Chemistry, 18, 2012
|
|
4F7R
 
 | |
5LU1
 
 | | Human 14-3-3 sigma CLU3 mutant complexed with short HSPB6 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heat shock protein beta-6 | | Authors: | Sluchanko, N.N, Beelen, S, Kulikova, A.A, Weeks, S.D, Antson, A.A, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Structure, 25, 2017
|
|
5LX2
 
 | |
5LU2
 
 | | Human 14-3-3 sigma complexed with long HSPB6 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Heat shock protein beta-6 | | Authors: | Sluchanko, N.N, Beelen, S, Kulikova, A.A, Weeks, S.D, Antson, A.A, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Structure, 25, 2017
|
|
5MHC
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, LYS-LEU-MET-PHE-LYS-TPO-GLU-GLY-PRO-ASP-SER-ASP, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-11-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
5MOC
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
5MYC
 
 | | Crystal structure of human 14-3-3 sigma in complex with LRRK2 peptide pS910 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stevers, L.M, de Vries, R.M.J.M, Ottmann, C. | | Deposit date: | 2017-01-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structural interface between LRRK2 and 14-3-3 protein.
Biochem. J., 474, 2017
|
|
5MXO
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide stabilized by Fusicoccin-A | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, FUSICOCCIN, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
5M37
 
 | | The molecular tweezer CLR01 stabilizes a disordered protein-protein interface | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein zeta/delta, BENZOIC ACID, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Molecular Tweezer CLR01 Stabilizes a Disordered Protein-Protein Interface.
J. Am. Chem. Soc., 139, 2017
|
|
5MY9
 
 | | Crystal structure of human 14-3-3 sigma in complex with LRRK2 peptide pS935 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stevers, L.M, de Vries, R.M.J.M, Ottmann, C. | | Deposit date: | 2017-01-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.327 Å) | | Cite: | Structural interface between LRRK2 and 14-3-3 protein.
Biochem. J., 474, 2017
|
|
5LVZ
 
 | | Crystal structure of yeast 14-3-3 protein from Lachancea thermotolerans | | Descriptor: | KLTH0G14146p | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of a yeast 14-3-3 protein from Lachancea thermotolerans in the unliganded form and bound to a human lipid kinase PI4KB-derived peptide reveal high evolutionary conservation.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5N5T
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV2 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyphenol, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
