8K6K
 
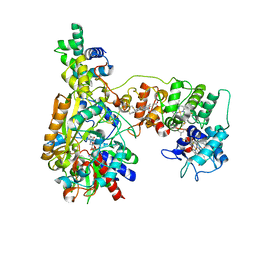 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (N1146A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
7WSQ
 
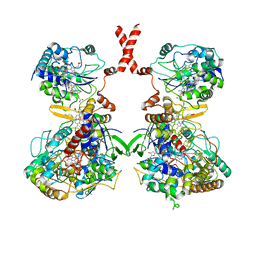 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
7VZG
 
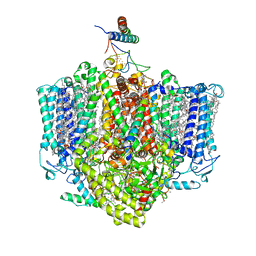 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
8OVD
 
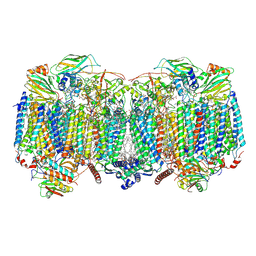 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
8OVC
 
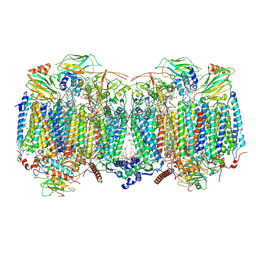 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
7VZR
 
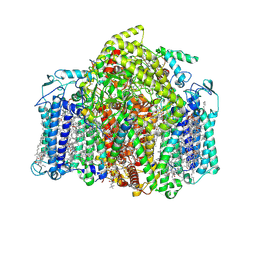 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
7W2J
 
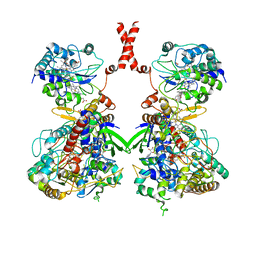 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Makino, F, Miyata, T, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry
Acs Catalysis, 13, 2023
|
|
4NFG
 
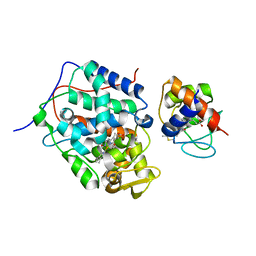 | | K13R mutant of horse cytochrome c and yeast cytochrome c peroxidase complex | | Descriptor: | Cytochrome c, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Meulenbroek, E.M, Bashir, Q, Ubbink, M, Pannu, N.S. | | Deposit date: | 2013-10-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Engineering specificity in a dynamic protein complex with a single conserved mutation.
Febs J., 281, 2014
|
|
3VRD
 
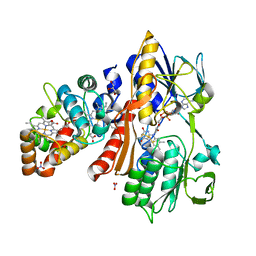 | | Crystal structure of flavocytochrome c from Thermochromatium tepidum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavocytochrome c flavin subunit, Flavocytochrome c heme subunit, ... | | Authors: | Hirano, Y, Kimura, Y, Suzuki, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure Analysis and Comparative Characterization of the Cytochrome c' and Flavocytochrome c from Thermophilic Purple Photosynthetic Bacterium Thermochromatium tepidum
Biochemistry, 51, 2012
|
|
7E1V
 
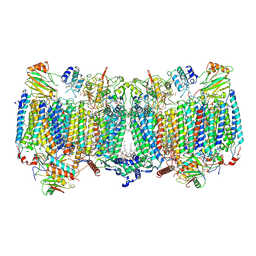 | | Cryo-EM structure of apo hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, CARDIOLIPIN, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-13 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
7E1X
 
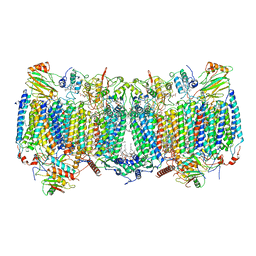 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in presence of TB47 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 5-methoxy-2-methyl-~{N}-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]pyrazolo[1,5-a]pyridine-3-carboxamide, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
7E1W
 
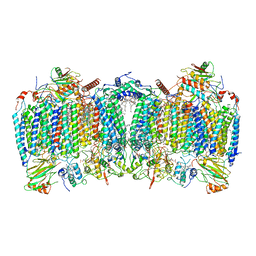 | | Cryo-EM structure of hybrid respiratory supercomplex consisting of Mycobacterium tuberculosis complexIII and Mycobacterium smegmatis complexIV in the presence of Q203 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, 6-chloranyl-2-ethyl-N-[[4-[4-[4-(trifluoromethyloxy)phenyl]piperidin-1-yl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide, ... | | Authors: | Zhou, S, Wang, W, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Elife, 10, 2021
|
|
4P4Q
 
 | |
3WFE
 
 | | Reduced and cyanide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CYANIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
5N1T
 
 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | Descriptor: | COPPER (II) ION, CopC, Cytochrome C, ... | | Authors: | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
3HQ6
 
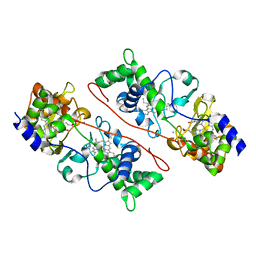 | | Cytochrome c peroxidase from G. sulfurreducens, wild type | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
5OCF
 
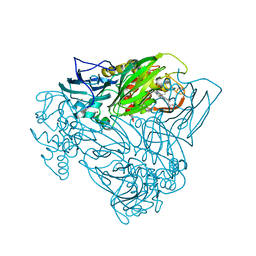 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OBO
 
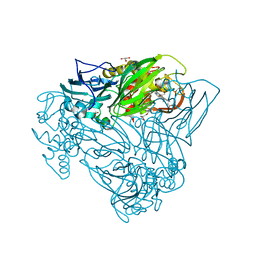 | | Crystal structure of nitrite bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, GLYCEROL, HEME C, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-28 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
3HQ8
 
 | | CcpA from G. sulfurreducens S134P/V135K variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, IMIDAZOLE, ... | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3HQ7
 
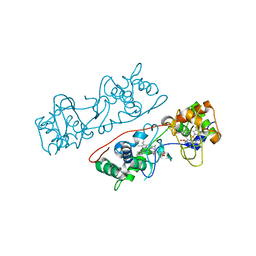 | | CcpA from G. sulfurreducens, G94K/K97Q/R100I variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3HQ9
 
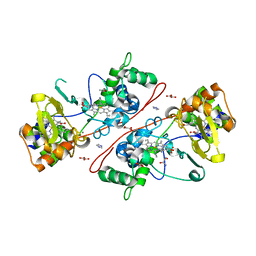 | | CcpA from G. sulfurreducens, S134P variant | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, Cytochrome c551 peroxidase, ... | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
3J2T
 
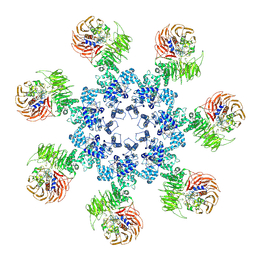 | | An improved model of the human apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Yuan, S, Topf, M, Akey, C.W. | | Deposit date: | 2012-12-23 | | Release date: | 2013-04-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Changes in apaf-1 conformation that drive apoptosome assembly.
Biochemistry, 52, 2013
|
|
