6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EF3
 
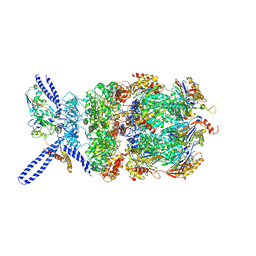 | | Yeast 26S proteasome bound to ubiquitinated substrate (4D motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EM8
 
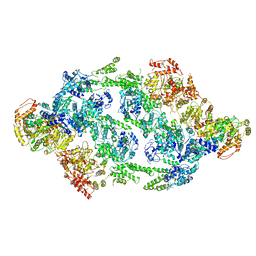 | | S.aureus ClpC resting state, C2 symmetrised | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC | | Authors: | Carroni, M, Mogk, A. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6EMW
 
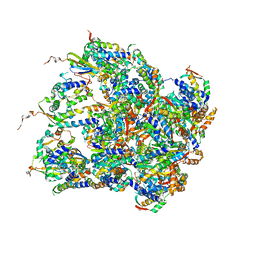 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6F0X
 
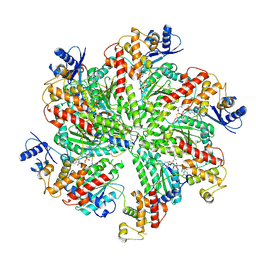 | | Cryo-EM structure of TRIP13 in complex with ATP gamma S, p31comet, C-Mad2 and Cdc20 | | Descriptor: | Cell division cycle protein 20 homolog, MAD2L1-binding protein, Mitotic spindle assembly checkpoint protein MAD2A, ... | | Authors: | Alfieri, C, Chang, L, Barford, D. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13.
Nature, 559, 2018
|
|
6FVU
 
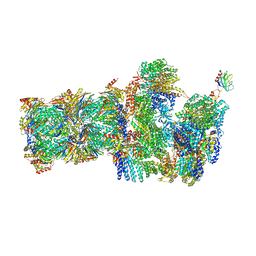 | | 26S proteasome, s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6FVX
 
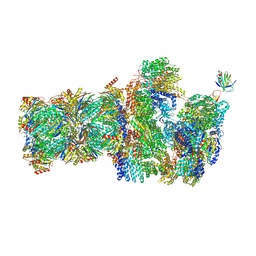 | | 26S proteasome, s5 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6GCO
 
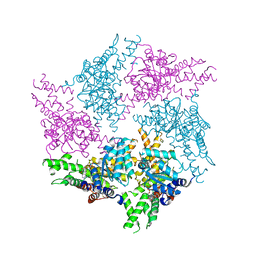 | | Truncated FtsH from A. aeolicus in P312 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, ZINC ION | | Authors: | Uthoff, M, Baumann, U. | | Deposit date: | 2018-04-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Conformational flexibility of pore loop-1 gives insights into substrate translocation by the AAA+protease FtsH.
J. Struct. Biol., 204, 2018
|
|
6G2V
 
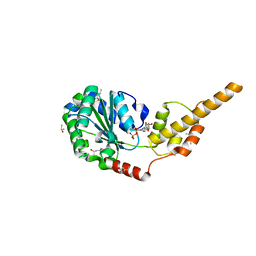 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2W
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.678 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6FVW
 
 | | 26S proteasome, s4 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6G2Y
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2Z
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6GCN
 
 | | Truncated FtsH from A. aeolicus in R32 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, ATP-dependent zinc metalloprotease FtsH, ... | | Authors: | Uthoff, M, Baumann, U. | | Deposit date: | 2018-04-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Conformational flexibility of pore loop-1 gives insights into substrate translocation by the AAA+protease FtsH.
J. Struct. Biol., 204, 2018
|
|
6FVY
 
 | | 26S proteasome, s6 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6G30
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.418 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6FVV
 
 | | 26S proteasome, s3 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6G2X
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6FVT
 
 | | 26S proteasome, s1 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6P11
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with JNJ-7706621 inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6P10
 
 | | Structure of spastin AAA domain (N527C mutant) in complex with JNJ-7706621 inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
6PE0
 
 | | Msp1 (E214Q)-substrate complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Membrane-spanning ATPase-like protein, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
8RHN
 
 | | Structure of the 55LCC ATPase complex | | Descriptor: | ATPase family gene 2 protein homolog A, ATPase family gene 2 protein homolog B, Cyclin-dependent kinase 2-interacting protein, ... | | Authors: | Foglizzo, M, Degtjarik, O, Zeqiraj, E. | | Deposit date: | 2023-12-15 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
