7FBC
 
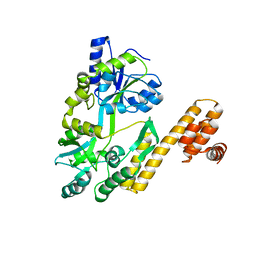 | | De novo design protein D22 with MBP tag | | Descriptor: | Maltodextrin-binding protein,De novo design protein D22 | | Authors: | Bin, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
1LAX
 
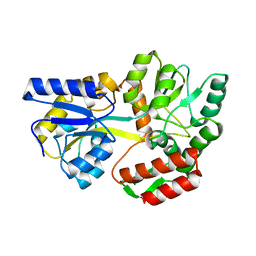 | | CRYSTAL STRUCTURE OF MALE31, A DEFECTIVE FOLDING MUTANT OF MALTOSE-BINDING PROTEIN | | Descriptor: | MALTOSE-BINDING PROTEIN MUTANT MALE31, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Mourez, M, Vulliez-le Normand, B, Sassoon, N, Bentley, G.A, Betton, J.M. | | Deposit date: | 2002-03-29 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a defective folding protein
PROTEIN SCI., 12, 2003
|
|
3SEY
 
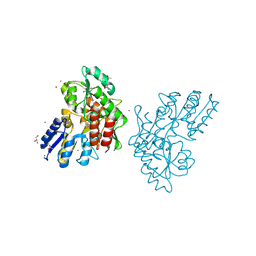 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
5VX2
 
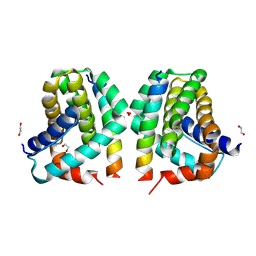 | | Mcl-1 in complex with Bim-h3Pc-RT | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1 chimera | | Authors: | Cowan, A.D, Brouwer, J.M, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
3RUM
 
 | | New strategy to analyze structures of glycopeptide antibiotic-target complexes | | Descriptor: | 3-amino-2,3,6-trideoxy-alpha-L-ribo-hexopyranose, ISOPROPYL ALCOHOL, Maltose-binding periplasmic protein, ... | | Authors: | Economou, N.J, Weeks, S.D, Grasty, K.C, Nahoum, V, Loll, P.J. | | Deposit date: | 2011-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A carrier protein strategy yields the structure of dalbavancin.
J.Am.Chem.Soc., 134, 2012
|
|
5AQ9
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, OFF7_DB08V4 | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
3G7V
 
 | | Islet Amyloid Polypeptide (IAPP or Amylin) fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3WAI
 
 | |
4TSM
 
 | | MBP-fusion protein of PilA1 from C. difficile R20291 residues 26-166 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein, pilin chimera, ... | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-06-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
1MDQ
 
 | |
6QZ6
 
 | | Structure of Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4WMW
 
 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WVI
 
 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with a substrate peptide (pep2). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, substrate peptide (pep2) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
4WMS
 
 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
1EZ9
 
 | |
1JVY
 
 | | Maltodextrin-binding protein variant D207C/A301GS/P316C with beta-mercaptoethanol mixed disulfides | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltodextrin-binding protein | | Authors: | Srinivasan, U, Iyer, G.H, Przybycien, T.A, Samsonoff, W.A, Bell, J.A. | | Deposit date: | 2001-08-31 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystine: Fibrous Biomolecular Material from Protein Crystals Cross-linked in a Specific Geometry
Protein Eng., 15, 2002
|
|
7NB4
 
 | | Structure of Mcl-1 complex with compound 1 | | Descriptor: | (2~{R})-2-[[5-(3-chloranyl-2-methyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Core Replacement on S64315, a Selective MCL-1 Inhibitor, and Its Analogues.
Acs Omega, 6, 2021
|
|
3WIX
 
 | | Crystal structure of Mcl-1 in complex with compound 4 | | Descriptor: | 7-(4-carboxyphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
1FQB
 
 | | STRUCTURE OF MALTOTRIOTOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
3SES
 
 | | Cu-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SET
 
 | | Ni-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form I) | | Descriptor: | Maltose-binding periplasmic protein, NICKEL (II) ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
8AX6
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0029882 | | Descriptor: | (1~{S},10~{R},20~{E})-12-methyl-10-[(7-methyl-2~{H}-indazol-5-yl)methyl]-15,18-dioxa-9,12,24,26-tetrazapentacyclo[20.5.2.1^{1,4}.1^{3,7}.0^{25,28}]hentriaconta-3(30),4,6,20,22,24,28-heptaene-8,11,27-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel Macrocyclic Antagonists of the CGRP Receptor Part 2: Stereochemical Inversion Induces an Unprecedented Binding Mode.
Acs Med.Chem.Lett., 13, 2022
|
|
1FQA
 
 | | STRUCTURE OF MALTOTETRAITOL BOUND TO OPEN-FORM MALTODEXTRIN BINDING PROTEIN IN P2(1)CRYSTAL FORM | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-sorbitol | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
6QB3
 
 | | Apo Mcl1 in a complex with a scFv | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Kazmirski, S, Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
