8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
6DBQ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3J9P
 
 | | Structure of the TRPA1 ion channel determined by electron cryo-microscopy | | Descriptor: | Maltose-binding periplasmic protein, Transient receptor potential cation channel subfamily A member 1 chimera | | Authors: | Paulsen, C.E, Armache, J.-P, Gao, Y, Cheng, Y, Julius, D. | | Deposit date: | 2015-02-14 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structure of the TRPA1 ion channel suggests regulatory mechanisms.
Nature, 520, 2015
|
|
6DBV
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.291 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBT
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7TT6
 
 | | BamABCDE bound to substrate EspP in the intermediate-open EspP state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT3
 
 | | BamABCDE bound to substrate EspP class 5 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT1
 
 | | BamABCDE bound to substrate EspP class 4 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT0
 
 | | BamABCDE bound to substrate EspP class 2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT5
 
 | | BamABCDE bound to substrate EspP in the open-sheet EspP state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
6DBO
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of substrate RSS DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7TSZ
 
 | | BamABCDE bound to substrate EspP class 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7BY0
 
 | | The cryo-EM structure of CENP-A nucleosome in complex with the phosphorylated CENP-C | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
8CRB
 
 | | Cryo-EM structure of PcrV/Fab(11-E5) | | Descriptor: | Heavy chain, Light chain, Maltose/maltodextrin-binding periplasmic protein,Type III secretion protein PcrV | | Authors: | Yuan, B, Simonis, A, Marlovits, T.C. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Discovery of highly neutralizing human antibodies targeting Pseudomonas aeruginosa.
Cell, 186, 2023
|
|
6DBW
 
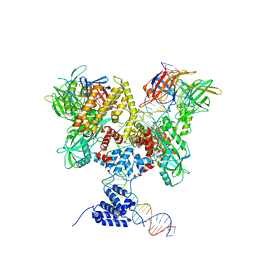 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7TT7
 
 | | BamABCDE bound to substrate EspP in the barrelized EspP/continuous open BamA state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
6DBL
 
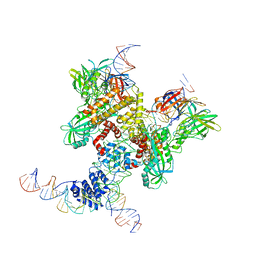 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.001 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7JIJ
 
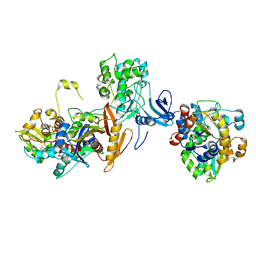 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7RJ5
 
 | | The structure of BAM in complex with EspP at 7 Angstrom resolution | | Descriptor: | Maltodextrin-binding protein,Autotransporter outer membrane beta-barrel domain-containing protein chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
8T2I
 
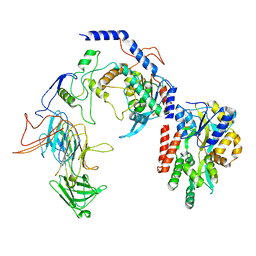 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | Descriptor: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | Authors: | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
5ZR0
 
 | | Solution structure of peptidyl-prolyl cis/trans isomerase domain of Trigger Factor in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein,Trigger factor | | Authors: | Kawagoe, S, Nakagawa, H, Kumeta, H, Ishimori, K, Saio, T. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into prolinecis/transisomerization of unfolded proteins catalyzed by the trigger factor chaperone.
J. Biol. Chem., 293, 2018
|
|
1EZO
 
 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
1EZP
 
 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN USING PEPTIDE ORIENTATIONS FROM DIPOLAR COUPLINGS | | Descriptor: | MALTODEXTRIN BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
2V93
 
 | | EQUILLIBRIUM MIXTURE OF OPEN AND PARTIALLY-CLOSED SPECIES IN THE APO STATE OF MALTODEXTRIN-BINDING PROTEIN BY PARAMAGNETIC RELAXATION ENHANCEMENT NMR | | Descriptor: | 1-(1-HYDROXY-2,2,6,6-TETRAMETHYLPIPERIDIN-4-YL)PYRROLIDINE-2,5-DIONE, MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Clore, G.M, Tang, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR.
Nature, 449, 2007
|
|
