6W6K
 
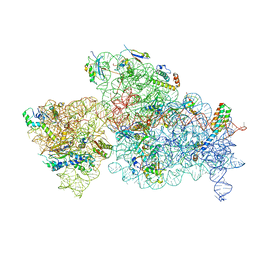 | | 30S-Activated-high-Mg2+ | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
5JTE
 
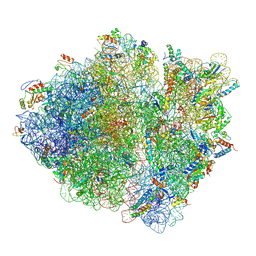 | | Cryo-EM structure of an ErmBL-stalled ribosome in complex with A-, P-, and E-tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Bock, L.V, Graf, M, Innis, C.A, Beckmann, R, Grubmueller, H, Vaiana, A.C, Wilson, D.N. | | Deposit date: | 2016-05-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest.
Nat Commun, 7, 2016
|
|
6W77
 
 | | 30S-Inactivated-high-Mg2+ Class A | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
5KCR
 
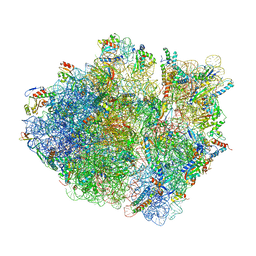 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5UYL
 
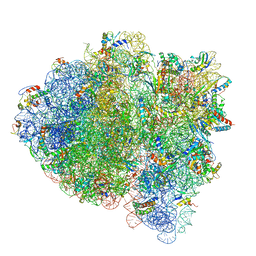 | | 70S ribosome bound with cognate ternary complex base-paired to A site codon (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity.
Nature, 546, 2017
|
|
3JBU
 
 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | Deposit date: | 2015-10-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
8PHJ
 
 | | cA4-bound Cami1 in complex with 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA (2862-MER), 5S rRNA, ... | | Authors: | Tamulaitiene, G, Mogila, I, Sasnauskas, G, Tamulaitis, G. | | Deposit date: | 2023-06-20 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Ribosomal stalk-captured CARF-RelE ribonuclease inhibits translation following CRISPR signaling.
Science, 382, 2023
|
|
6WDJ
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure V-A1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
5L3P
 
 | |
6WD6
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure II-C2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WD4
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure II-B2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WDL
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure V-B1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WD8
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure III-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WD9
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure III-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6XDQ
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6Q9A
 
 | |
6ENJ
 
 | |
4V89
 
 | | Crystal Structure of Release Factor RF3 Trapped in the GTP State on a Rotated Conformation of the Ribosome (without viomycin) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Trakhanov, S, Noller, H.F. | | Deposit date: | 2011-11-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome.
Rna, 18, 2012
|
|
6OUO
 
 | | RF2 accommodated state bound 70S complex at long incubation time | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCJ
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
7JSS
 
 | |
7JT3
 
 | |
7JSZ
 
 | |
6VU3
 
 | | Cryo-EM structure of Escherichia coli transcription-translation complex A (TTC-A) containing mRNA with a 12 nt long spacer | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
