7VN5
 
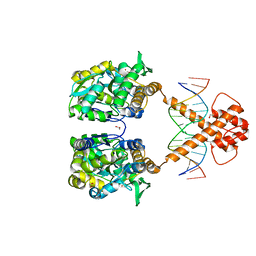 | | Crystal structure of MBP-fused BIL1/BZR1 (21-90) in complex with double-stranded DNA contaning TTCACGTGAA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*AP*A)-3'), Maltodextrin-binding protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Tanokura, M, Miyakawa, T. | | Deposit date: | 2021-10-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Brassinosteroid-induced gene repression requires specific and tight promoter binding of BIL1/BZR1 via DNA shape readout.
Nat.Plants, 8, 2022
|
|
5T0A
 
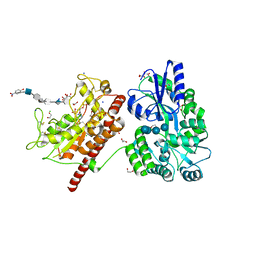 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5BJZ
 
 | |
4KYC
 
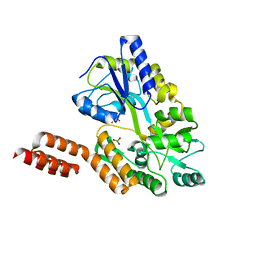 | | Structure of the C-terminal domain of the Menangle virus phosphoprotein, fused to MBP. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, Maltose-binding periplasmic protein, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
3C4M
 
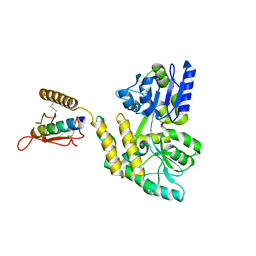 | |
5T05
 
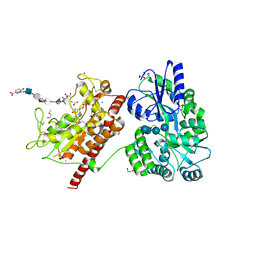 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and IdoA2S containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5GRU
 
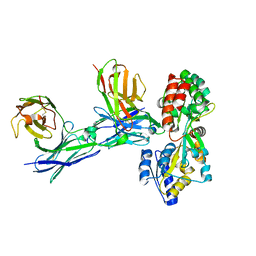 | | Structure of mono-specific diabody | | Descriptor: | Maltose-binding periplasmic protein, diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5AZ7
 
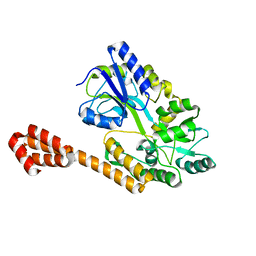 | |
3EHU
 
 | |
4XA2
 
 | |
4EGC
 
 | |
4IFP
 
 | | X-ray Crystal Structure of Human NLRP1 CARD Domain | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Jin, T, Curry, J, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1.
Proteins, 81, 2013
|
|
3CSB
 
 | |
1JW4
 
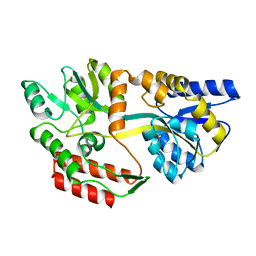 | | Structure of ligand-free maltodextrin-binding protein | | Descriptor: | maltodextrin-binding protein | | Authors: | Duan, X, Quiocho, F.A. | | Deposit date: | 2001-09-02 | | Release date: | 2002-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands.
Biochemistry, 41, 2002
|
|
4R0Y
 
 | |
1JW5
 
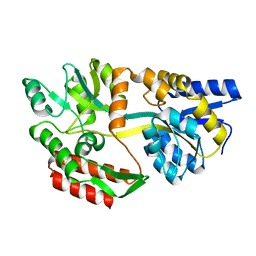 | |
5GPP
 
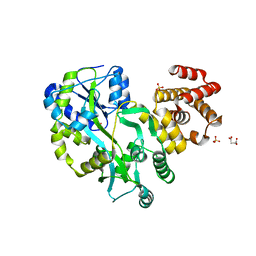 | | Crystal structure of zebrafish ASC PYD domain | | Descriptor: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
3LC8
 
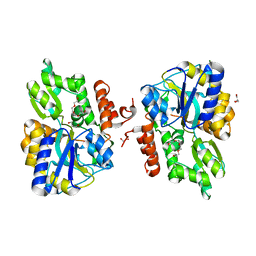 | |
7OMT
 
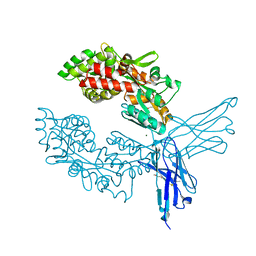 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
1MPB
 
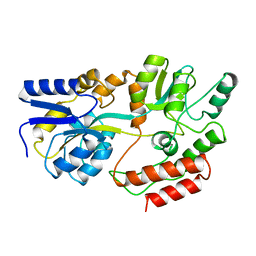 | |
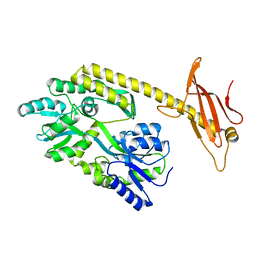
6K7D
 
 | |
4WMX
 
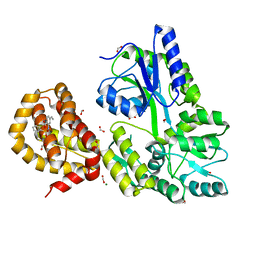 | | The structure of MBP-MCL1 bound to ligand 6 at 2.0A | | Descriptor: | 4-ethenyl-2-[(phenylsulfonyl)amino]benzoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
6KI0
 
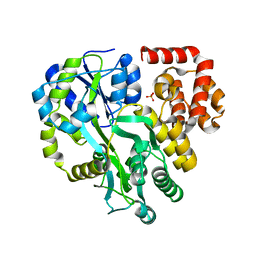 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
3HPI
 
 | |
1ZIU
 
 | |
