2VPG
 
 | | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | | Descriptor: | B-CELL CLL/LYMPHOMA 9 PROTEIN, GLYCEROL, HISTONE H3 TAIL, ... | | Authors: | Fiedler, M, Sanchez-Barrena, M.J, Nekrasov, M, Mieszczanek, J, Rybin, V, Muller, J, Bienz, M, Evans, P. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Mol.Cell, 30, 2008
|
|
4Y6L
 
 | | Human SIRT2 in complex with myristoylated peptide (H3K9myr) | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, peptide THR-ALA-ARG-MYK-SER-THR-GLY | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
5T1I
 
 | | CBX3 chromo shadow domain in complex with histone H3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
J. Biol. Chem., 292, 2017
|
|
4LK9
 
 | | Crystal Structure of MOZ double PHD finger histone H3 tail complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
5U2J
 
 | |
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
4LKA
 
 | | Crystal Structure of MOZ double PHD finger histone H3K9ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
1Q3L
 
 | |
5JHN
 
 | | Structure of G9a SET-domain with Histone H3K9Ala mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9A mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LUG
 
 | | Crystal structure of human Spindlin-2B protein in complex with ART(M3L)QTA(2MR)KS peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-M3L-GLN-THR-ALA-2MR-LYS-SER, N3, ... | | Authors: | Srikannathasan, V, Gileadi, C, Talon, R, Shrestha, L, Kopec, J, Szykowska, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Oppermann, U, Huber, K. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Spindlin-2B protein in complex with ART(M3L)QTA(2MR)KS peptide
To be published
|
|
5T0K
 
 | | Structure of G9a SET-domain with H3K9M mutant peptide and SAM | | Descriptor: | H3K9 mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading.
Elife, 5, 2016
|
|
8HMX
 
 | |
5TDW
 
 | | Set3 PHD finger in complex with histone H3K4me3 | | Descriptor: | SET domain-containing protein 3, SODIUM ION, ZINC ION, ... | | Authors: | Andrews, F.H, Ali, M, Kutateladze, T.G. | | Deposit date: | 2016-09-19 | | Release date: | 2016-10-19 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
J. Mol. Biol., 429, 2017
|
|
6WAV
 
 | | Crystal structure of PHF1 in complex with H3K36me3 substitution | | Descriptor: | Histone H3.1, PHD finger protein 1, SULFATE ION, ... | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
4F56
 
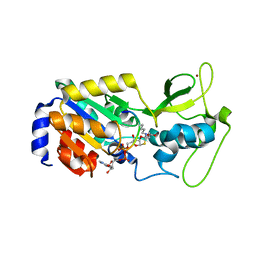 | | The bicyclic intermediate structure provides insights into the desuccinylation mechanism of SIRT5 | | Descriptor: | 3-[(2R,3aR,5R,6R,6aR)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-2,6-dihydroxytetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]propanoic acid, NAD-dependent lysine demalonylase and desuccinylase sirtuin-5, mitochondrial, ... | | Authors: | Zhou, Y, Hao, Q. | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5)
J.Biol.Chem., 287, 2012
|
|
5VAB
 
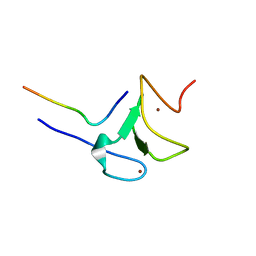 | | Crystal structure of ATXR5 PHD domain in complex with histone H3 | | Descriptor: | ATXR5 PHD domain, Histone H3 peptide, ZINC ION | | Authors: | Bergamin, E, Sarvan, S, Malette, J, Eram, M, Yeung, S, Mongeon, V, Joshi, M, Brunzelle, J.S, Michaels, S.D, Blais, A, Vedadi, M, Couture, J.-F. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5JJ0
 
 | | Structure of G9a SET-domain with Histone H3K9M peptide and excess SAH | | Descriptor: | Histone H3K9M mutant peptide, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7EBK
 
 | |
5MR8
 
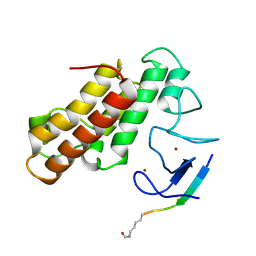 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide | | Descriptor: | E3 ubiquitin-protein ligase TRIM33, Histone H3, ZINC ION | | Authors: | Tallant, C, Savitsky, P, Fedorov, O, Nunez-Alonso, G, Siejka, P, Krojer, T, Williams, E, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide
To Be Published
|
|
6MIQ
 
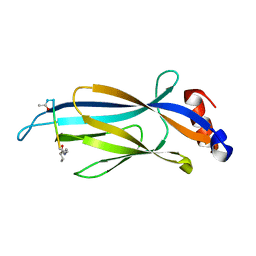 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | Descriptor: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | Authors: | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
4Z0R
 
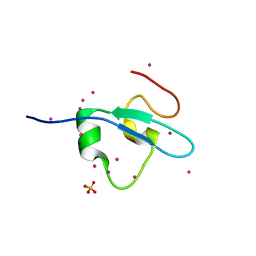 | | Crystal Structure of the CW domain of ZCWPW2 mutant F78R in complex with histone H3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Histone H3.1, SULFATE ION, ... | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the CW domain of ZCWPW2 mutant F78R in complex with histone H3 peptide
To be Published
|
|
1O9S
 
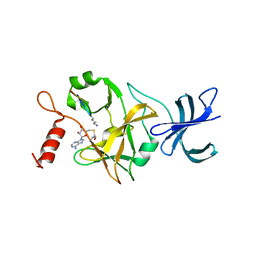 | | Crystal structure of a ternary complex of the human histone methyltransferase SET7/9 | | Descriptor: | GENE FRAGMENT FOR HISTONE H3, HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC, ... | | Authors: | Xiao, B, Jing, C, Wilson, J.R, Walker, P.A, Vasisht, N, Kelly, G, Howell, S, Taylor, I.A, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Human Histone Methyltransferase Set7/9
Nature, 421, 2003
|
|
3O35
 
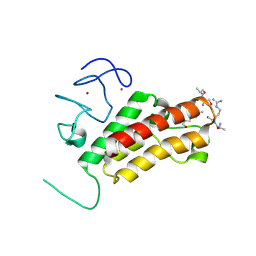 | |
2VNF
 
 | | MOLECULAR BASIS OF HISTONE H3K4ME3 RECOGNITION BY ING4 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, HISTONE H3, ... | | Authors: | Palacios, A, Munoz, I.G, Pantoja-Uceda, D, Marcaida, M.J, Torres, D, Martin-Garcia, J.M, Luque, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular Basis of Histone H3K4Me3 Recognition by Ing4
J.Biol.Chem., 283, 2008
|
|
