1HPC
 
 | | REFINED STRUCTURES AT 2 ANGSTROMS AND 2.2 ANGSTROMS OF THE TWO FORMS OF THE H-PROTEIN, A LIPOAMIDE-CONTAINING PROTEIN OF THE GLYCINE DECARBOXYLASE | | Descriptor: | 5-[(3S)-1,2-dithiolan-3-yl]pentanoic acid, H PROTEIN OF THE GLYCINE CLEAVAGE SYSTEM, LIPOIC ACID | | Authors: | Pares, S, Cohen-Addad, C, Sieker, L, Neuburger, M, Douce, R. | | Deposit date: | 1994-02-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures at 2 and 2.2 A resolution of two forms of the H-protein, a lipoamide-containing protein of the glycine decarboxylase complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1HPG
 
 | |
1HPH
 
 | |
1HPI
 
 | | MOLECULAR STRUCTURE OF THE OXIDIZED HIGH-POTENTIAL IRON-SULFUR PROTEIN ISOLATED FROM ECTOTHIORHODOSPIRA VACUOLATA | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Benning, M.M, Meyer, T.E, Rayment, I, Holden, H.M. | | Deposit date: | 1993-12-09 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structure of the oxidized high-potential iron-sulfur protein isolated from Ectothiorhodospira vacuolata.
Biochemistry, 33, 1994
|
|
1HPJ
 
 | |
1HPK
 
 | |
1HPL
 
 | |
1HPM
 
 | |
1HPN
 
 | | N.M.R. AND MOLECULAR-MODELLING STUDIES OF THE SOLUTION CONFORMATION OF HEPARIN | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Mulloy, B, Forster, M.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | N.m.r. and molecular-modelling studies of the solution conformation of heparin.
Biochem.J., 293, 1993
|
|
1HPO
 
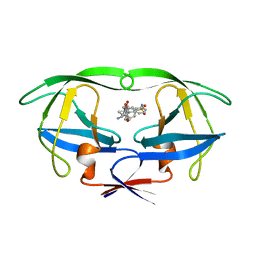 | | HIV-1 PROTEASE TRIPLE MUTANT/U103265 COMPLEX | | Descriptor: | 4-CYANO-N-(3-CYCLOPROPYL(5,6,7,8,9,10-HEXAHYDRO-4-HYDROXY-2-OXO-CYCLOOCTA[B]PYRAN-3-YL)METHYL)PHENYL BENZENSULFONAMIDE, HIV-1 PROTEASE | | Authors: | Watenpaugh, K.D, Janakiraman, M.N. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design of nonpeptidic HIV protease inhibitors: the sulfonamide-substituted cyclooctylpyramones.
J.Med.Chem., 40, 1997
|
|
1HPS
 
 | | RATIONAL DESIGN, SYNTHESIS AND CRYSTALLOGRAPHIC ANALYSIS OF A HYDROXYETHYLENE-BASED HIV-1 PROTEASE INHIBITOR CONTAINING A HETEROCYCLIC P1'-P2' AMIDE BOND ISOSTERE | | Descriptor: | 2-[(1R,3S,4S)-1-BENZYL-4-[N-(BENZYLOXYCARBONYL)-L-VALYL]AMINO-3-PHENYLPENTYL]-4(5)-(2-METHYLPROPIONYL)IMIDAZOLE, HIV-1 PROTEASE | | Authors: | Abdel-Meguid, S, Zhao, B. | | Deposit date: | 1994-05-24 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis, and crystallographic analysis of a hydroxyethylene-based HIV-1 protease inhibitor containing a heterocyclic P1'--P2' amide bond isostere.
J.Med.Chem., 37, 1994
|
|
1HPT
 
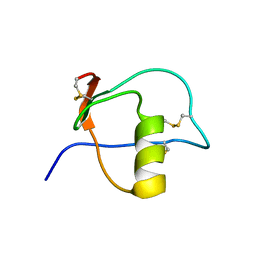 | |
1HPU
 
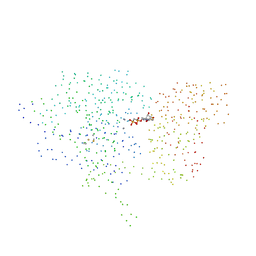 | | 5'-NUCLEOTIDASE (CLOSED FORM), COMPLEX WITH AMPCP | | Descriptor: | 5'-NUCLEOTIDASE, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-13 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
1HPV
 
 | |
1HPW
 
 | | STRUCTURE OF A PILIN MONOMER FROM PSEUDOMONAS AERUGINOSA: IMPLICATIONS FOR THE ASSEMBLY OF PILI. | | Descriptor: | FIMBRIAL PROTEIN | | Authors: | Keizer, D.W, Slupsky, C.M, Campbell, A.P, Irvin, R.T, Sykes, B.D. | | Deposit date: | 2000-12-13 | | Release date: | 2001-05-02 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a pilin monomer from Pseudomonas aeruginosa: implications for the assembly of pili.
J.Biol.Chem., 276, 2001
|
|
1HPX
 
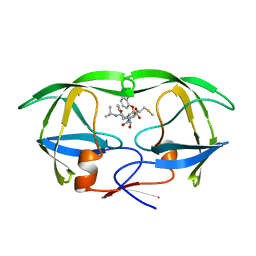 | | HIV-1 PROTEASE COMPLEXED WITH THE INHIBITOR KNI-272 | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, HIV-1 PROTEASE | | Authors: | Bhat, T.N, Erickson, J.W. | | Deposit date: | 1995-05-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine.
Structure, 3, 1995
|
|
1HPY
 
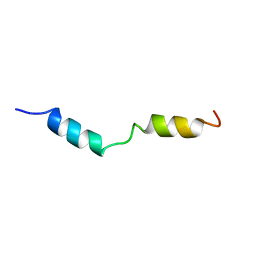 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-34 IN 20% TRIFLUORETHANOL, NMR, 10 STRUCTURES | | Descriptor: | PARATHYROID HORMONE | | Authors: | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | Deposit date: | 1998-09-30 | | Release date: | 2000-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
1HPZ
 
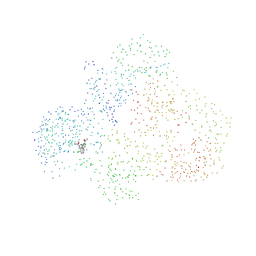 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
1HQ0
 
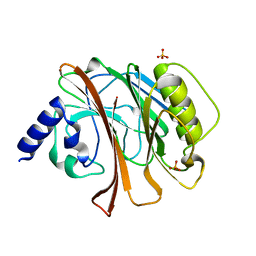 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF E.COLI CYTOTOXIC NECROTIZING FACTOR TYPE 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2000-12-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
1HQ1
 
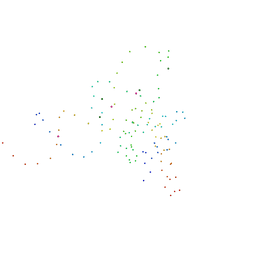 | | STRUCTURAL AND ENERGETIC ANALYSIS OF RNA RECOGNITION BY A UNIVERSALLY CONSERVED PROTEIN FROM THE SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Sagar, M.B, Doudna, J.A. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle.
J.Mol.Biol., 307, 2001
|
|
1HQ2
 
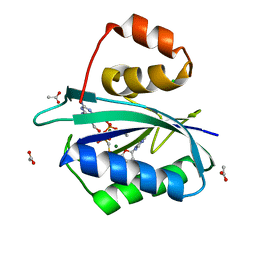 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF E.COLI HPPK(R82A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.25 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ACETATE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-12-13 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HQ4
 
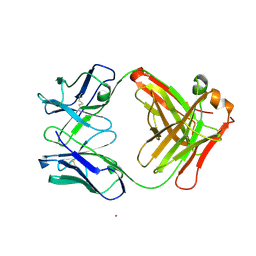 | | STRUCTURE OF NATIVE CATALYTIC ANTIBODY HA5-19A4 | | Descriptor: | ANTIBODY HA5-19A4 FAB HEAVY CHAIN, ANTIBODY HA5-19A4 FAB LIGHT CHAIN, CADMIUM ION | | Authors: | Paschall, C.M, Hasserodt, J, Lerner, R.A, Janda, K.D, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polyene Cyclization Reactions
Thesis, 2000
|
|
1HQ5
 
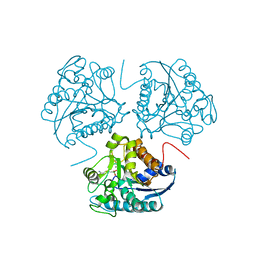 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH S-(2-BORONOETHYL)-L-CYSTEINE, AN L-ARGININE ANALOGUE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Kim, N.N, Cox, J.D, Baggio, R.F, Emig, F.A, Mistry, S.K, Harper, S.L, Speicher, D.W, Morris Jr, S.M, Ash, D.E, Traish, A, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
Biochemistry, 40, 2001
|
|
1HQ6
 
 | | STRUCTURE OF PYRUVOYL-DEPENDENT HISTIDINE DECARBOXYLASE AT PH 8 | | Descriptor: | HISTIDINE DECARBOXYLASE | | Authors: | Schelp, E, Worley, S, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | pH-induced structural changes regulate histidine decarboxylase activity in Lactobacillus 30a.
J.Mol.Biol., 306, 2001
|
|
