5DAY
 
 | | The structure of NAP1-Related Protein(NRP1) in Arabidopsis | | Descriptor: | CALCIUM ION, NAP1-related protein 1 | | Authors: | Zhu, Y, Rong, L, Yang, Y, Zhang, C, Feng, H.Y, Zheng, L.N, Shen, W.H, Ma, J.B, Dong, A.W. | | Deposit date: | 2015-08-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | The structure of NAP1-Related Protein(NRP1) in Arabidopsis
To Be Published
|
|
3QFN
 
 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH in complex with inorganic phosphate | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
6ODC
 
 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6ODA
 
 | | Crystal structure of HDAC8 in complex with compound 2 | | Descriptor: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
7N44
 
 | | Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13) | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Reilly, R.A, Zhang, C.H, Deshmukh, M.G, Ippolito, J.A, Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency.
Acs Med.Chem.Lett., 12, 2021
|
|
5ZIU
 
 | | Crystal structure of human Entervirus D68 RdRp | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, RdRp | | Authors: | Wang, M.L, Zhang, Y, Chen, Y.P, Lu, D.R, Jiang, H, Chen, Y.J, Li, L, Zhang, C.H, Shi, Q.L, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Crystal structure of human Entervirus D68 RdRp in complex with NADPH
To Be Published
|
|
1ZXF
 
 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
3E6U
 
 | | Crystal structure of Human LanCL1 | | Descriptor: | LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
3E73
 
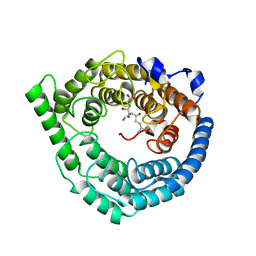 | | Crystal Structure of Human LanCL1 complexed with GSH | | Descriptor: | GLUTATHIONE, LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | Descriptor: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,Probable G-protein coupled receptor 34,YL-365 | | Authors: | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3D9H
 
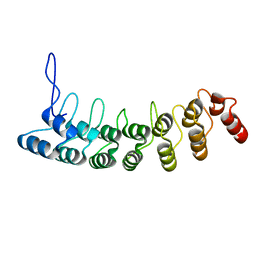 | | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein | | Descriptor: | cDNA FLJ77766, highly similar to Homo sapiens ankyrin repeat and SOCS box-containing 9 (ASB9), transcript variant 2, ... | | Authors: | Fei, X, Gu, X, Fan, S, Zhang, C, Ji, C. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein
To be published
|
|
5L6P
 
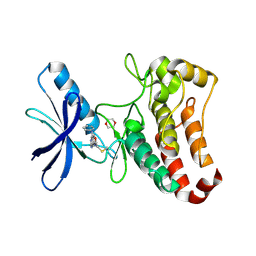 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 6) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ephrin type-B receptor 3, ~{N}-(4-phenylazanylquinazolin-7-yl)ethanamide | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
5L6O
 
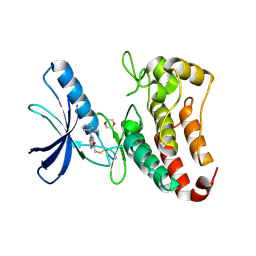 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 3) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 1-(4-phenylazanylquinazolin-7-yl)ethanone, Ephrin type-B receptor 3 | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
1YXU
 
 | | Crystal Structure of Kinase Pim1 in Complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structures of Proto-oncogene Kinase Pim1: A Target of Aberrant Somatic Hypermutations in Diffuse Large Cell Lymphoma.
J.Mol.Biol., 348, 2005
|
|
5XK4
 
 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XK5
 
 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1ZDT
 
 | | The Crystal Structure of Human Steroidogenic Factor-1 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, Steroidogenic factor 1 | | Authors: | Wang, W, Zhang, C, Marimuthu, A, Krupka, H.I, Tabrizizad, M, Shelloe, R, Mehra, U, Eng, K, Nguyen, H, Settachatgul, C, Powell, B, Milburn, M.V, West, B.L. | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6D26
 
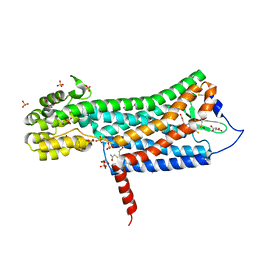 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
8JYS
 
 | |
6D27
 
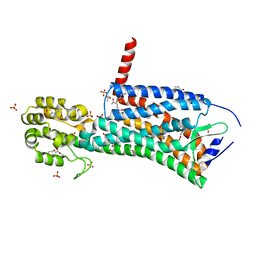 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6C1Q
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, PMX53, Soluble cytochrome b562, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
