8HCS
 
 | | zebrafish IRF-11 DBD | | Descriptor: | Interferon regulatory factor | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-03 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
3T2K
 
 | | Crystal structure of sulfide:quinone oxidoreductase Cys128Ala variant from Acidithiobacillus ferrooxidans with bound trisulfane | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-22 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
8JFQ
 
 | | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-back Loop | | Descriptor: | 26mer-DNA | | Authors: | Liu, Y, Li, J, Zhang, Y, Wang, Y, Chen, J, Bian, Y, Xia, Y, Yang, M.H, Zheng, K, Wang, K.B, Kong, L.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop.
J.Am.Chem.Soc., 145, 2023
|
|
7Y35
 
 | | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex
To Be Published
|
|
7Y36
 
 | | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HYDROXIDE ION, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-10 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex
to be published
|
|
3T2Y
 
 | | Crystal structure of sulfide:quinone oxidoreductase His132Ala variant from Acidithiobacillus ferrooxidans with bound disulfide | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-23 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
6KJ6
 
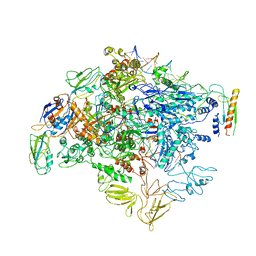 | | cryo-EM structure of Escherichia coli Crl transcription activation complex | | Descriptor: | DNA (51-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Xu, J, Zhang, Y. | | Deposit date: | 2019-07-21 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor.
Elife, 8, 2019
|
|
7DTK
 
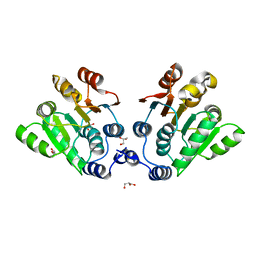 | |
6J9F
 
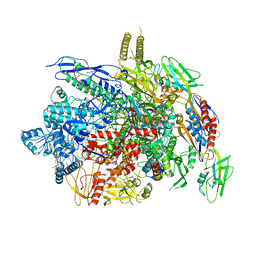 | |
3TKJ
 
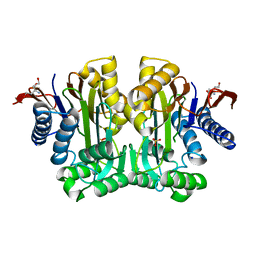 | | Crystal Structure of Human Asparaginase-like Protein 1 Thr168Ala | | Descriptor: | L-asparaginase, SODIUM ION, SULFATE ION, ... | | Authors: | Li, W.Z, Yogesha, S.D, Liu, J, Zhang, Y. | | Deposit date: | 2011-08-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncoupling Intramolecular Processing and Substrate Hydrolysis in the N-Terminal Nucleophile Hydrolase hASRGL1 by Circular Permutation.
Acs Chem.Biol., 7, 2012
|
|
8H9F
 
 | | Human ATP synthase state 1 subregion 3 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
7EH2
 
 | |
7EH1
 
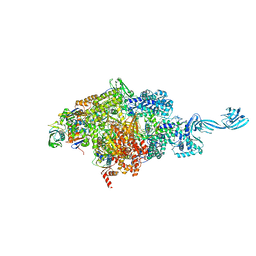 | | Thermus thermophilus transcription initiation complex containing a template-strand purine at position TSS-2, GpG RNA primer, and CMPcPP | | Descriptor: | 1,4-BUTANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (27-MER), ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E1Z
 
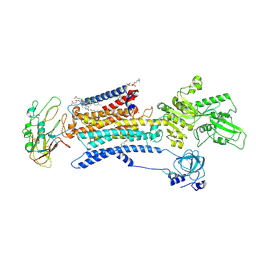 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E20
 
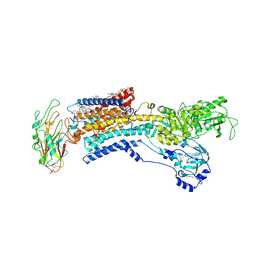 | | Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E21
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
8IOF
 
 | |
8ID1
 
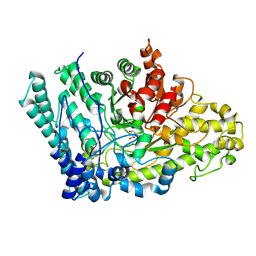 | |
8IMZ
 
 | | Cryo-EM structure of mouse Piezo1-MDFIC complex (composite map) | | Descriptor: | MyoD family inhibitor domain-containing protein, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhou, Z, Ma, X, Lin, Y, Cheng, D, Bavi, N, Li, J.V, Sutton, D, Yao, M, Harvey, N, Corry, B, Zhang, Y, Cox, C.D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | MyoD-family inhibitor proteins act as auxiliary subunits of Piezo channels.
Science, 381, 2023
|
|
7DWY
 
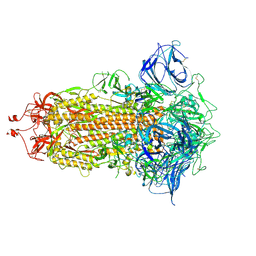 | | S protein of SARS-CoV-2 in the locked conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX6
 
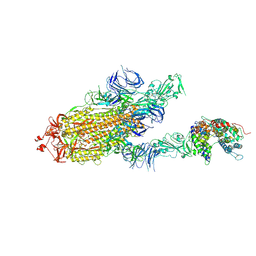 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 3 (2 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DWX
 
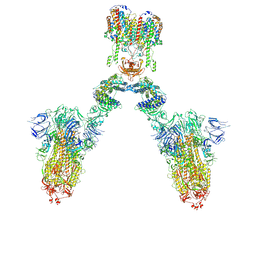 | | Conformation 1 of S-ACE2-B0AT1 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX3
 
 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 1 (1 up RBD and no PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX0
 
 | | Trypsin-digested S protein of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX1
 
 | | S protein of SARS-CoV-2 D614G mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
