2DLR
 
 | |
2DLV
 
 | |
2CRP
 
 | |
2CRW
 
 | |
6M10
 
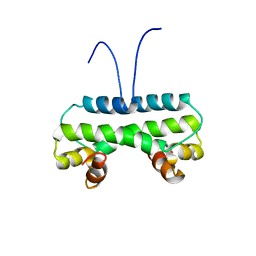 | | Crystal structure of PA4853 (Fis) from Pseudomonas aeruginosa | | Descriptor: | Putative Fis-like DNA-binding protein | | Authors: | Zhang, H, Gao, Z, Zhou, J, Dong, Y. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of the nucleoid-associated protein Fis (PA4853) from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2YSR
 
 | |
3EUO
 
 | |
8JGA
 
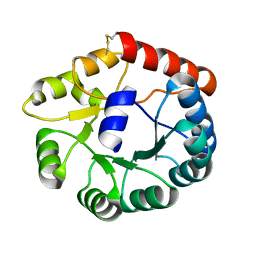 | | Cryo-EM structure of Mi3 fused with FKBP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8JGC
 
 | | Cryo-EM structure of Mi3 fused with LOV2 | | Descriptor: | LOV domain-containing protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4F0V
 
 | |
4F0W
 
 | |
4F46
 
 | | Crystal structure of wild type human CD38 in complex with NAADP and ADPRP | | Descriptor: | ADP-ribosyl cyclase 1, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5S)-3,4,5-tris(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2012-05-10 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structures of Human CD38 in Complex with NAADP and ADPRP
MESSENGER, 2, 2013
|
|
5V4A
 
 | |
6O1T
 
 | | BOVINE SALIVARY PROTEIN FORM 30B WITH OLEIC ACID | | Descriptor: | CALCIUM ION, OLEIC ACID, Short palate, ... | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
4F45
 
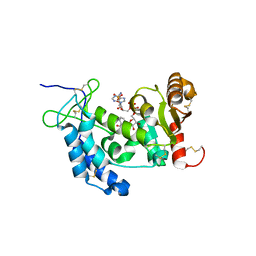 | | Crystal structure of human CD38 E226Q mutant in complex with NAADP | | Descriptor: | ADP-ribosyl cyclase 1, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2012-05-10 | | Release date: | 2013-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human CD38 in Complex with NAADP and ADPRP
MESSENGER, 2, 2013
|
|
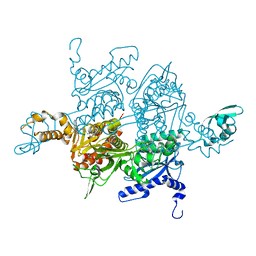
3H0J
 
 | |
2OW3
 
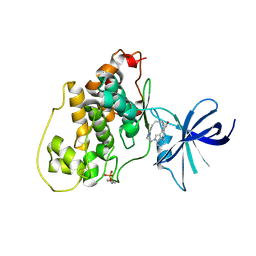 | | Glycogen synthase kinase-3 beta in complex with bis-(indole)maleimide pyridinophane inhibitor | | Descriptor: | BIS-(INDOLE)MALEIMIDE PYRIDINOPHANE, Glycogen synthase kinase-3 beta | | Authors: | Zhang, H.C, Bonaga, L.V, Ye, H, Derian, C.K, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2007-02-15 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel bis(indolyl)maleimide pyridinophanes that are potent, selective inhibitors of glycogen synthase kinase-3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3HFJ
 
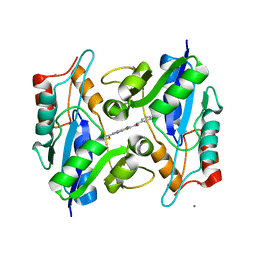 | | Bacillus anthracis nicotinate mononucleotide adenylytransferase (nadD) in complex with inhibitor CID 3289443 | | Descriptor: | 3-amino-N-(3-fluorophenyl)-6-thiophen-2-ylthieno[2,3-b]pyridine-2-carboxamide, CALCIUM ION, Nicotinate (Nicotinamide) nucleotide adenylyltransferase | | Authors: | Zhang, H, Huang, N, Eyobo, Y. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Targeting NAD biosynthesis in bacterial pathogens: Structure-based development of inhibitors of nicotinate mononucleotide adenylyltransferase NadD.
Chem.Biol., 16, 2009
|
|
6NIG
 
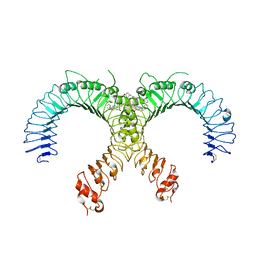 | | Crystal structure of the human TLR2-Diprovocim complex | | Descriptor: | (3S,4S,3'S,4'S)-1,1'-(1,4-phenylenedicarbonyl)bis{N~3~,N~4~-bis[(1S,2R)-2-phenylcyclopropyl]pyrrolidine-3,4-dicarboxami de}, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Beutler, B.A, Tomchick, D.R, Su, L. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim.
J. Med. Chem., 62, 2019
|
|
8FK2
 
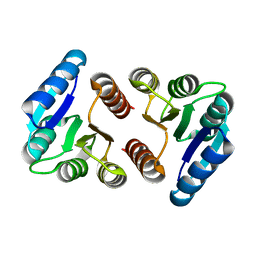 | | The N-terminal VicR from Streptococcus mutans | | Descriptor: | Putative response regulator CovR VicR-like protein | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Small Molecule Attenuates Bacterial Virulence by Targeting Conserved Response Regulator.
Mbio, 14, 2023
|
|
6ITM
 
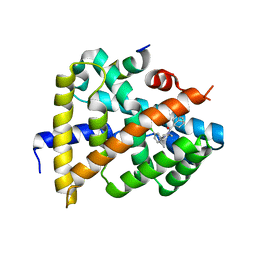 | | Crystal structure of FXR in complex with agonist XJ034 | | Descriptor: | 1-adamantyl-[4-(5-chloranyl-2-methyl-phenyl)piperazin-1-yl]methanone, Bile acid receptor, HD3 Peptide from Nuclear receptor coactivator 1 | | Authors: | Zhang, H, Wang, Z. | | Deposit date: | 2018-11-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pose Filter-Based Ensemble Learning Enables Discovery of Orally Active, Nonsteroidal Farnesoid X Receptor Agonists.
J.Chem.Inf.Model., 60, 2020
|
|
5V2A
 
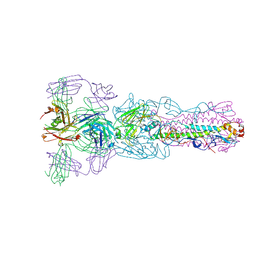 | | Crystal structure of Fab H7.167 in complex with influenza virus hemagglutinin from A/Shanghai/02/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H7.167 antibody, Hemagglutinin, ... | | Authors: | Zhang, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.656 Å) | | Cite: | H7N9 influenza virus neutralizing antibodies that possess few somatic mutations.
J. Clin. Invest., 126, 2016
|
|
7DW5
 
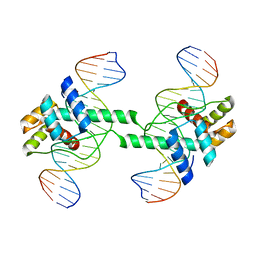 | | Crystal structure of DUX4 HD1-HD2 domain complexed with ERG sites | | Descriptor: | BROMIDE ION, DNA (5'-D(P*CP*GP*AP*CP*TP*TP*GP*AP*TP*GP*AP*GP*AP*TP*TP*AP*GP*AP*CP*TP*G)-3'), Double homeobox protein 4-like protein 2 | | Authors: | Zhang, H, Cheng, N, Li, Z, Zhang, W, Dong, X, Huang, J, Meng, G. | | Deposit date: | 2021-01-15 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Cancer Commun (Lond), 41, 2021
|
|
7EJ3
 
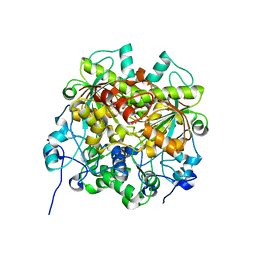 | | UTP cyclohydrolase | | Descriptor: | DIPHOSPHATE, GLYCINE, GTP cyclohydrolase II, ... | | Authors: | Zhang, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-04-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical investigation of UTP cyclohydrolase
Acs Catalysis, 2021
|
|
