6NSF
 
 | |
3C2A
 
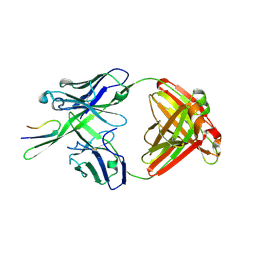 | | Antibody Fab fragment 447-52D in complex with UG1033 peptide | | Descriptor: | Envelope glycoprotein, Fab 447-52D heavy chain, Fab 447-52D light chain | | Authors: | Dhillon, A.K, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2008-01-24 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6O30
 
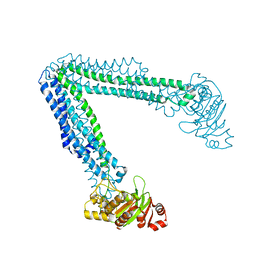 | | Lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Zhang, Q, Wilson, I.A, Lee, S.C, Stanfield, R.L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.47 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
6O3D
 
 | |
6O3J
 
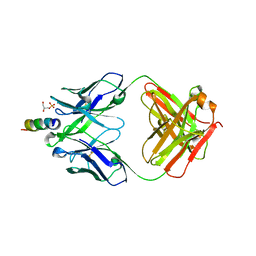 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.416 Å) | | Cite: | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
6O3L
 
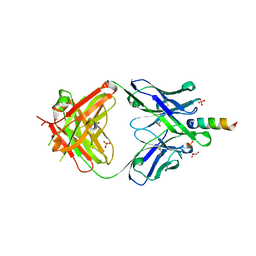 | |
6O41
 
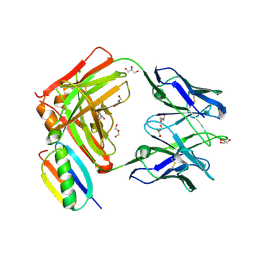 | |
3CFJ
 
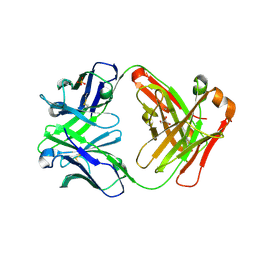 | |
6OPA
 
 | |
6O42
 
 | |
6O3K
 
 | |
6O3U
 
 | |
6OO0
 
 | | Crystal structure of bovine Fab NC-Cow1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NC-Cow1 heavy chain, NC-Cow1 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of broad HIV neutralization by a vaccine-induced cow antibody.
Sci Adv, 6, 2020
|
|
3CN3
 
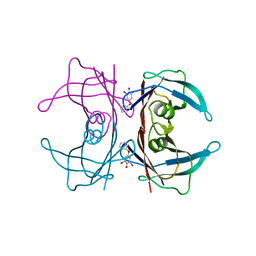 | |
6OC7
 
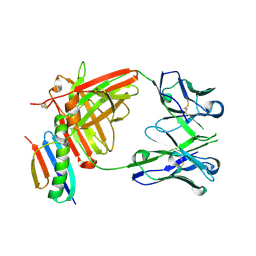 | | HMP42 Fab in complex with Protein G | | Descriptor: | Heavy chain of HMP42 Fab, Immunoglobulin G-binding protein G, Light chain for HMP42 Fab | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2019-03-22 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
3CFC
 
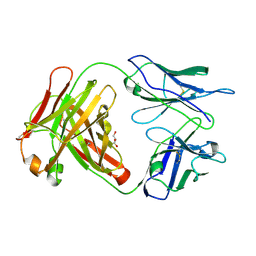 | | High-resolution structure of blue fluorescent antibody EP2-19G2 | | Descriptor: | BLUE FLUORESCENT ANTIBODY EP2-19G2-IGG2B HEAVY CHAIN, BLUE FLUORESCENT ANTIBODY EP2-19G2-KAPPA LIGHT CHAIN, GLYCEROL | | Authors: | Debler, E.W, Wilson, I.A. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deeply inverted electron-hole recombination in a luminescent antibody-stilbene complex.
Science, 319, 2008
|
|
3CN0
 
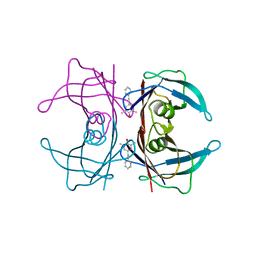 | |
3CN4
 
 | |
3BEQ
 
 | | Neuraminidase of A/Brevig Mission/1/1918 H1N1 strain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, X, Zhu, X, Wilson, I.A. | | Deposit date: | 2007-11-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of the 1918 influenza virus H1N1 neuraminidase
J.Virol., 82, 2008
|
|
3CN1
 
 | |
1IAO
 
 | | CLASS II MHC I-AD IN COMPLEX WITH OVALBUMIN PEPTIDE 323-339 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II I-AD | | Authors: | Scott, C.A, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 1998-03-13 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues.
Immunity, 8, 1998
|
|
6NSA
 
 | |
6NSG
 
 | |
3B7E
 
 | | Neuraminidase of A/Brevig Mission/1/1918 H1N1 strain in complex with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Xu, X, Zhu, X, Wilson, I.A. | | Deposit date: | 2007-10-30 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural characterization of the 1918 influenza virus H1N1 neuraminidase
J.Virol., 82, 2008
|
|
6PBW
 
 | | Crystal structure of Fab667 complex | | Descriptor: | Fab667 heavy chain, Fab667 light chain, GLYCEROL, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.058 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
