5DFZ
 
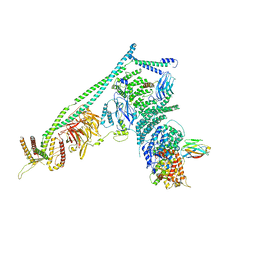 | | Structure of Vps34 complex II from S. cerevisiae. | | Descriptor: | Nanobody binding S. cerevisiae Vps34, Phosphatidylinositol 3-kinase VPS34, Putative N-terminal domain of S. cerevisiae Vps30, ... | | Authors: | Rostislavleva, K, Soler, N, Ohashi, Y, Zhang, L, Williams, R.L. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science, 350, 2015
|
|
1E7U
 
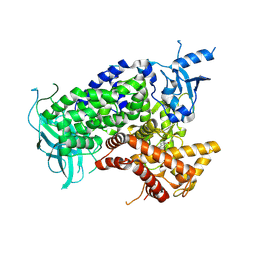 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Perisic, O, Ried, C, Stephens, L, Williams, R.L. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E8W
 
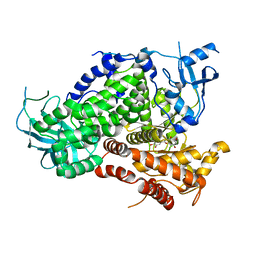 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E8Y
 
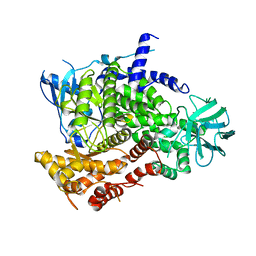 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E8Z
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT, STAUROSPORINE | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1E90
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1DJY
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-2,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DJZ
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-4,5-BISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-4,5-BISPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1NHK
 
 | |
1OEY
 
 | | Heterodimer of p40phox and p67phox PB1 domains from human NADPH oxidase | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 2, NEUTROPHIL CYTOSOL FACTOR 4 | | Authors: | Wilson, M.I, Gill, D.J, Perisic, O, Quinn, M.T, Williams, R.L. | | Deposit date: | 2003-04-02 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pb1 Domain-Mediated Heterodimerization in Nadph Oxidase and Signaling Complexes of Atypical Protein Kinase C with Par6 and P62
Mol.Cell, 12, 2003
|
|
1E7V
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine.
Mol.Cell, 6, 2000
|
|
1NLI
 
 | | Complex of [E160A-E189A] trichosanthin and adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein alpha-trichosanthin | | Authors: | Shaw, P.C, Wong, K.B, Chan, D.S.B, Williams, R.L. | | Deposit date: | 2003-01-07 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for the interaction of [E160A-E189A]-trichosanthin with adenine.
Toxicon, 41, 2003
|
|
1DJX
 
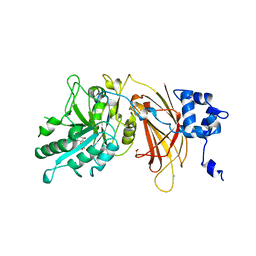 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-1,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1O7K
 
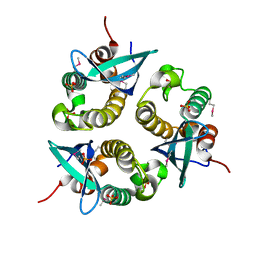 | | human p47 PX domain complex with sulphates | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 1, SULFATE ION | | Authors: | Karathanassis, D, Bravo, J, Perisic, O, Pacold, C.M, Williams, R.L. | | Deposit date: | 2002-11-07 | | Release date: | 2002-11-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of the Px Domain of P47Phox to Phosphatidylinositol 3.4-Bisphosphate and Phosphatidic Acid is Masked by an Intramolecular Interaction
Embo J., 21, 2002
|
|
1E8X
 
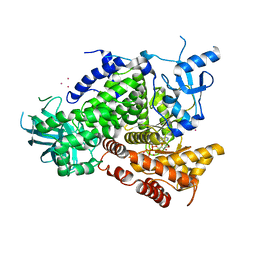 | | STRUCTURAL INSIGHTS INTO PHOSHOINOSITIDE 3-KINASE ENZYMATIC MECHANISM AND SIGNALLING | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LUTETIUM (III) ION, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Perisic, O, Ried, C, Stephens, L, Williams, R.L. | | Deposit date: | 2000-10-03 | | Release date: | 2000-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1DJW
 
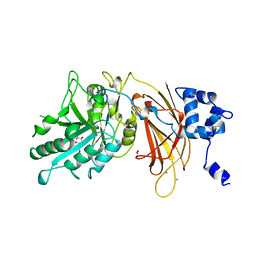 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHONATE | | Descriptor: | ACETATE ION, CALCIUM ION, INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
4CB8
 
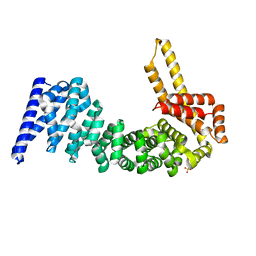 | | Structural and mutational analysis reveals that CTNNBL1 binds NLSs in a manner distinct from that of its closest armadillo-relative, karyopherin alpha | | Descriptor: | BETA-CATENIN-LIKE PROTEIN 1, SULFATE ION | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Mutational Analysis Reveals that Ctnnbl1 Binds Nlss in a Manner Distinct from that of its Closest Armadillo-Relative, Karyopherin Alpha
FEBS Lett., 588, 2014
|
|
4CBA
 
 | | Structural of delta 1-76 CTNNBL1 in space group I222 | | Descriptor: | 1,2-ETHANEDIOL, BETA-CATENIN-LIKE PROTEIN 1, SULFATE ION | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and mutational analysis reveals that CTNNBL1 binds NLSs in a manner distinct from that of its closest armadillo-relative, karyopherin alpha.
Febs Lett., 588, 2014
|
|
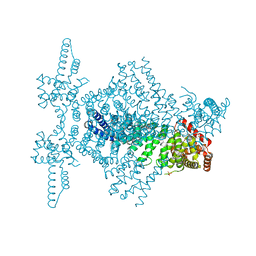
8RCK
 
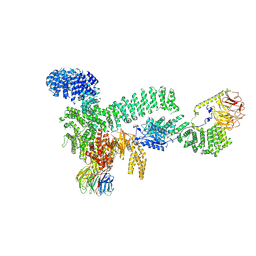 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy. | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy.
To Be Published
|
|
4A55
 
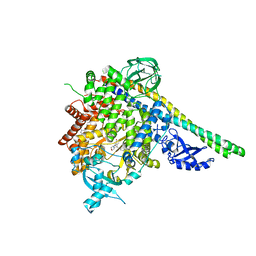 | | Crystal structure of p110alpha in complex with iSH2 of p85alpha and the inhibitor PIK-108 | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT ALPHA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM, PIK-108 | | Authors: | Hon, W.-C, Berndt, A, Williams, R.L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Regulation of Lipid Binding Underlies the Activation Mechanism of Class Ia Pi3-Kinases.
Oncogene, 31, 2012
|
|
4CB9
 
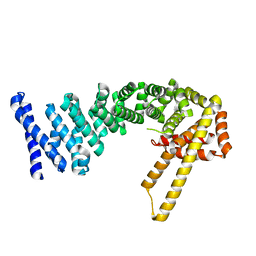 | | Structure of full-length CTNNBL1 in P43212 space group | | Descriptor: | BETA-CATENIN-LIKE PROTEIN 1 | | Authors: | Ganesh, K, vanMaldegem, F, Telerman, S.B, Simpson, P, Johnson, C.M, Williams, R.L, Neuberger, M.S, Rada, C. | | Deposit date: | 2013-10-10 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mutational Analysis Reveals that Ctnnbl1 Binds Nlss in a Manner Distinct from that of its Closest Armadillo-Relative, Karyopherin Alpha
FEBS Lett., 588, 2014
|
|
4AE4
 
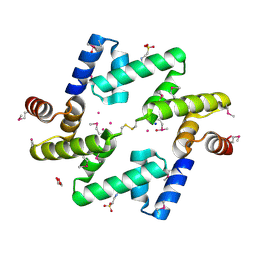 | | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a novel SOUBA domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Agromayor, M, Soler, N, Caballe, A, Kueck, T, Freund, S.M, Allen, M.D, Bycroft, M, Perisic, O, Ye, Y, McDonald, B, Scheel, H, Hofmann, K, Neil, S.J.D, Martin-Serrano, J, Williams, R.L. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-21 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain.
Structure, 20, 2012
|
|
4D0L
 
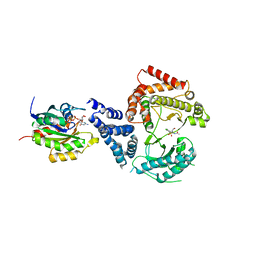 | | Phosphatidylinositol 4-kinase III beta-PIK93 in a complex with Rab11a- GTP gammaS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, ... | | Authors: | Burke, J.E, Inglis, A.J, Perisic, O, Masson, G.R, McLaughin, S.H, Rutaganira, F, Shokat, K.M, Williams, R.L. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Science, 344, 2014
|
|
4A5X
 
 | | Structures of MITD1 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, CHARGED MULTIVESICULAR BODY PROTEIN 1A, GLYCEROL, ... | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A5Z
 
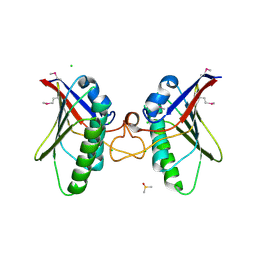 | | Structures of MITD1 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-30 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
