8J7I
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | crystal structure of SulE mutant
To Be Published
|
|
4KNF
 
 | | Crystal structure of a blue-light absorbing proteorhodopsin double-mutant D97N/Q105L from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KLY
 
 | | Crystal structure of a blue-light absorbing proteorhodopsin mutant D97N from HOT75 | | Descriptor: | Blue-light absorbing proteorhodopsin, RETINAL | | Authors: | Ran, T, Ozorowski, G, Gao, Y, Wang, W, Luecke, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4L4M
 
 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | Deposit date: | 2013-06-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
4L4L
 
 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 6-({4-[(3,5-difluorophenyl)sulfonyl]benzyl}carbamoyl)-1-(5-O-phosphono-beta-D-ribofuranosyl)imidazo[1,2-a]pyridin-1-ium, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | Deposit date: | 2013-06-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.122 Å) | | Cite: | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
8IVM
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVS
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7J
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW6
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVE
 
 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVN
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW3
 
 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVT
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7K
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7G
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
6B16
 
 | | P21-activated kinase 1 in complex with a 4-azaindole inhibitor | | Descriptor: | N~4~-(5-cyclopropyl-1H-pyrazol-3-yl)-N~2~-[(1S)-1-(1H-pyrrolo[3,2-b]pyridin-5-yl)ethyl]pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Rouge, L, Wang, W. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and evaluation of a series of 4-azaindole-containing p21-activated kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5T44
 
 | | Crystal structure of Frizzled 7 CRD | | Descriptor: | Frizzled-7 | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannous, R.H, Wang, W. | | Deposit date: | 2016-08-29 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.9944 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TUB
 
 | | Crystal Structure of the Shark TBC1D15 GAP Domain | | Descriptor: | Shark TBC1D15 GTPase-activating Protein | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
5TUC
 
 | | Crystal Structure of the Sus TBC1D15 GAP Domain | | Descriptor: | Sus TBC1D15 GAP Domain | | Authors: | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | Deposit date: | 2016-11-05 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
7EW4
 
 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW2
 
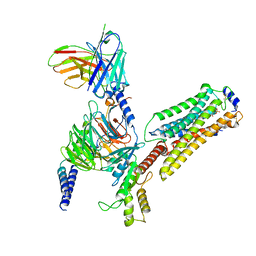 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW3
 
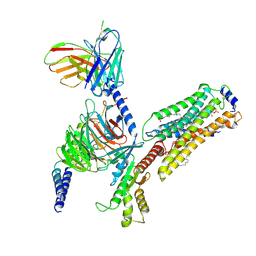 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
5URV
 
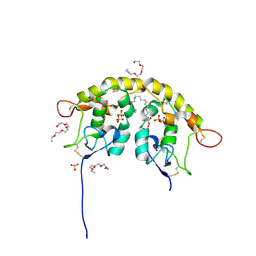 | | Crystal structure of Frizzled 7 CRD in complex with C24 fatty acid | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URZ
 
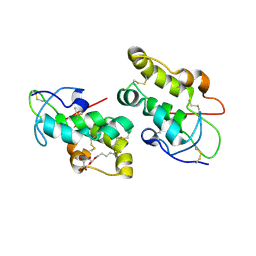 | | Crystal structure of Frizzled 5 CRD in complex with BOG | | Descriptor: | Frizzled-5, alpha-L-fucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, octyl beta-D-glucopyranoside | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URY
 
 | | Crystal structure of Frizzled 5 CRD in complex with PAM | | Descriptor: | Frizzled-5, PALMITOLEIC ACID, alpha-L-fucopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
