2LQU
 
 | |
2JT0
 
 | | Solution structure of F104W cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P.-J, Sykes, B.D. | | Deposit date: | 2007-07-17 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
6X13
 
 | | Inward-facing sodium-bound state of the glutamate transporter homologue GltPh | | Descriptor: | Glutamate transporter homologue GltPh, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, X, Boudker, O. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Large domain movements through the lipid bilayer mediate substrate release and inhibition of glutamate transporters.
Elife, 9, 2020
|
|
6X17
 
 | |
1JVJ
 
 | | CRYSTAL STRUCTURE OF N132A MUTANT OF TEM-1 BETA-LACTAMASE IN COMPLEX WITH A N-FORMIMIDOYL-THIENAMYCINE | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, BETA-LACTAMASE TEM, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2001-08-30 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Noncovalent interaction energies in covalent complexes: TEM-1 beta-lactamase and beta-lactams.
Proteins, 47, 2002
|
|
1JWZ
 
 | |
1JWV
 
 | |
6X12
 
 | | Inward-facing Apo-open state of the glutamate transporter homologue GltPh | | Descriptor: | Glutamate transporter homologue GltPh, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, X, Boudker, O. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Large domain movements through the lipid bilayer mediate substrate release and inhibition of glutamate transporters.
Elife, 9, 2020
|
|
6X14
 
 | | Inward-facing state of the glutamate transporter homologue GltPh in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, Glutamate transporter homologue GltPh, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, X, Boudker, O. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Large domain movements through the lipid bilayer mediate substrate release and inhibition of glutamate transporters.
Elife, 9, 2020
|
|
1JWP
 
 | |
1LHY
 
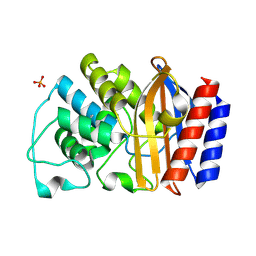 | | Crystal structure of TEM-30 beta-Lactamase at 2.0 Angstrom | | Descriptor: | Class A beta-Lactamase- TEM 30, PHOSPHATE ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
7YBJ
 
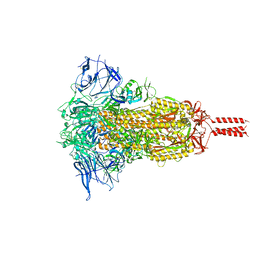 | | SARS-CoV-2 Mu variant spike(close state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBI
 
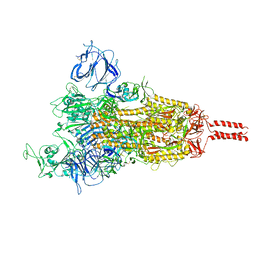 | | SARS-CoV-2 Mu variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
1LI0
 
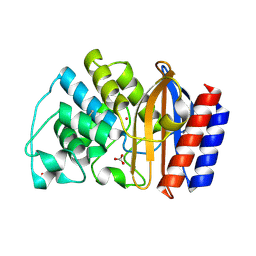 | | Crystal structure of TEM-32 beta-Lactamase at 1.6 Angstrom | | Descriptor: | BICARBONATE ION, Class A beta-Lactamase- TEM-32, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
7YBL
 
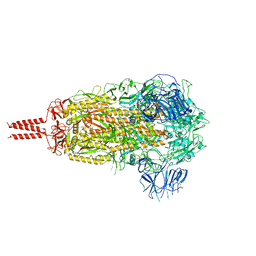 | | SARS-CoV-2 B.1.620 variant spike (close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBH
 
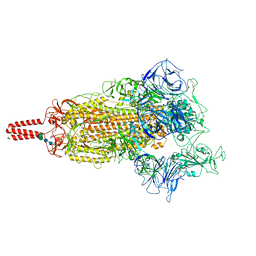 | | SARS-CoV-2 lambda variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBM
 
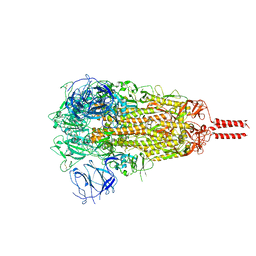 | | SARS-CoV-2 C.1.2 variant spike (Close state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBK
 
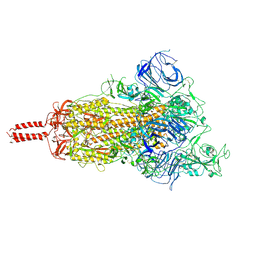 | | SARS-CoV-2 B.1.620 variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7YBN
 
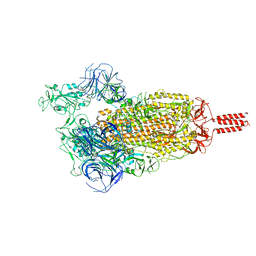 | | SARS-CoV-2 C.1.2 variant spike (Open state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
6X16
 
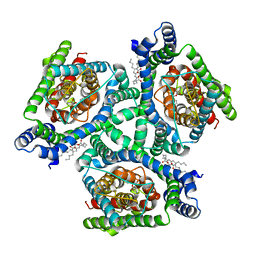 | | Inward-facing state of the glutamate transporter homologue GltPh in complex with TBOA | | Descriptor: | (3S)-3-(BENZYLOXY)-L-ASPARTIC ACID, Glutamate transporter homologue GltPh, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Wang, X, Boudker, O. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Large domain movements through the lipid bilayer mediate substrate release and inhibition of glutamate transporters.
Elife, 9, 2020
|
|
1LXF
 
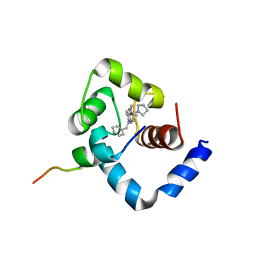 | | Structure of the Regulatory N-domain of Human Cardiac Troponin C in Complex with Human Cardiac Troponin-I(147-163) and Bepridil | | Descriptor: | 1-ISOBUTOXY-2-PYRROLIDINO-3[N-BENZYLANILINO] PROPANE, CALCIUM ION, TROPONIN C, ... | | Authors: | Wang, X, Li, M.X, Sykes, B.D. | | Deposit date: | 2002-06-05 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the regulatory N-domain of human
cardiac troponin C in complex with human cardiac
troponin I147-163 and bepridil.
J.Biol.Chem., 277, 2002
|
|
1LI9
 
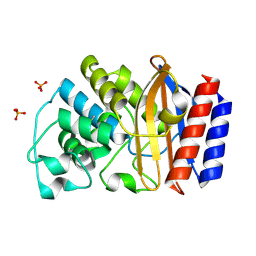 | | Crystal structure of TEM-34 beta-Lactamase at 1.5 Angstrom | | Descriptor: | Class A beta-Lactamase- TEM-34, PHOSPHATE ION, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
6L77
 
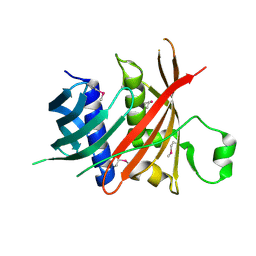 | | Crystal structure of MS5 from Brassica napus | | Descriptor: | MS5a | | Authors: | Wang, X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the meiosis-related protein MS5 reveals non-canonical papain enhancement by cystatin-like folds.
Febs Lett., 594, 2020
|
|
6M15
 
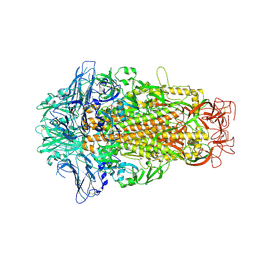 | | Cryo-EM structures of HKU2 spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
8WIL
 
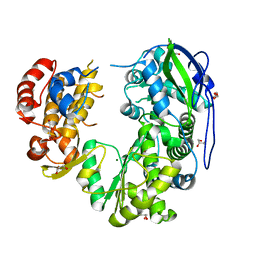 | | Crystal structure of Jingmen tick virus RNA-dependent RNA polymerase (D55 construct) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Jingmen tick virus NSP1, ... | | Authors: | Wang, X, Jing, X, Deng, F, Gong, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A jingmenvirus RNA-dependent RNA polymerase structurally resembles the flavivirus counterpart but with different features at the initiation phase.
Nucleic Acids Res., 52, 2024
|
|
