7EP9
 
 | |
2MVG
 
 | |
2MTC
 
 | |
2KJK
 
 | | Solution structure of the second domain of the listeria protein Lin2157, Northeast Structural Genomics Consortium target Lkr136b | | 分子名称: | Lin2157 protein | | 著者 | Wang, X, Hamilton, K, Xiao, R.H, Lee, D, Ciccosanti, C.H, Nair, R, Rost, B, Acton, T.B, Swapna, G, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-05-29 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Lkr136b
To be Published
|
|
7CN4
 
 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | 登録日 | 2020-07-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
2JZC
 
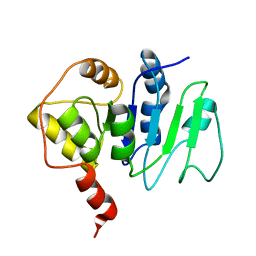 | | NMR solution structure of ALG13: The sugar donor subunit of a yeast N-acetylglucosamine transferase. Northeast Structural Genomics Consortium target YG1 | | 分子名称: | UDP-N-acetylglucosamine transferase subunit ALG13 | | 著者 | Wang, X, Weldeghorghis, T, Zhang, G, Imepriali, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-01-04 | | 公開日 | 2008-02-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase.
Structure, 16, 2008
|
|
7DH6
 
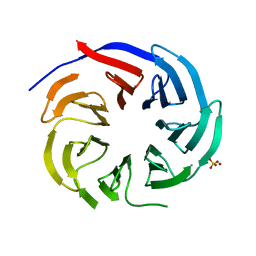 | | Crystal structure of PLRG1 | | 分子名称: | CALCIUM ION, NICKEL (II) ION, Pleiotropic regulator 1, ... | | 著者 | Wang, X, Xu, C. | | 登録日 | 2020-11-13 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.584 Å) | | 主引用文献 | Crystal structure of the WD40 domain of human PLRG1.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7CN8
 
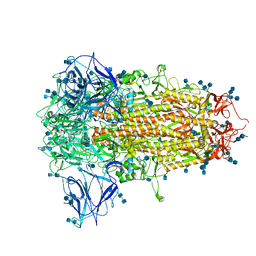 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | 著者 | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | 登録日 | 2020-07-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
2N77
 
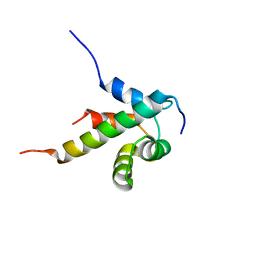 | |
2K7N
 
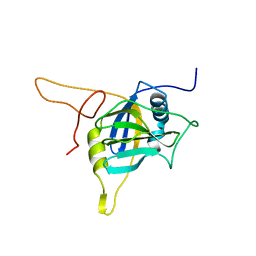 | |
7CYH
 
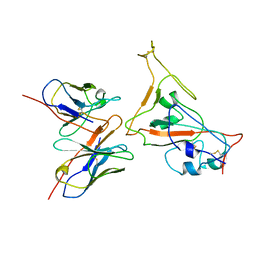 | |
2JT3
 
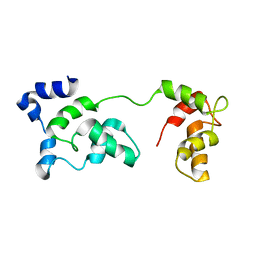 | | Solution Structure of F153W cardiac troponin C | | 分子名称: | Troponin C | | 著者 | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | 登録日 | 2007-07-18 | | 公開日 | 2007-07-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JTZ
 
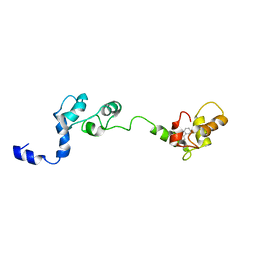 | | Solution structure and chemical shift assignments of the F104-to-5-flurotryptophan mutant of cardiac troponin C | | 分子名称: | Troponin C, slow skeletal and cardiac muscles | | 著者 | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | 登録日 | 2007-08-10 | | 公開日 | 2007-08-28 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
2JT0
 
 | | Solution structure of F104W cardiac troponin C | | 分子名称: | Troponin C, slow skeletal and cardiac muscles | | 著者 | Wang, X, Mercier, P, Letourneau, P.-J, Sykes, B.D. | | 登録日 | 2007-07-17 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JT8
 
 | | Solution structure of the F153-to-5-flurotryptophan mutant of human cardiac troponin C | | 分子名称: | Troponin C, slow skeletal and cardiac muscles | | 著者 | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | 登録日 | 2007-07-20 | | 公開日 | 2007-08-07 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
7BW0
 
 | | Active human TGR5 complex with a synthetic agonist 23H | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Chen, G, Wang, X.K, Chen, Q, Hu, H.L, Ren, R.B. | | 登録日 | 2020-04-12 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of activated bile acids receptor TGR5 in complex with stimulatory G protein.
Signal Transduct Target Ther, 5, 2020
|
|
8XRP
 
 | | The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, Interleukin-12 receptor subunit beta-2, ... | | 著者 | Chen, H.Q, Wang, X.Q, Ge, X.F, Zeng, J.W, Wang, J.W. | | 登録日 | 2024-01-07 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | The Cryo-EM structure of IL-12/IL-12RB1-D1/IL-12RB2-D1-D3 complex
To Be Published
|
|
6PFM
 
 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | 分子名称: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | 著者 | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | 登録日 | 2019-06-21 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2J14
 
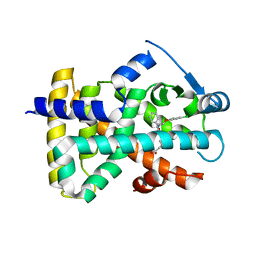 | | 3,4,5-Trisubstituted Isoxazoles as Novel PPARdelta Agonists: Part2 | | 分子名称: | (3-{4-[2-(2,4-DICHLORO-PHENOXY)-ETHYLCARBAMOYL]-5-PHENYL-ISOXAZOL-3-YL}-PHENYL)-ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR DELTA | | 著者 | Epple, R, Azimioara, M, Russo, R, Xie, Y, Wang, X, Cow, C, Wityak, J, Karanewsky, D, Bursulaya, B, Kreusch, A, Tuntland, T, Gerken, A, Iskandar, M, Saez, E, Seidel, H.M, Tian, S.S. | | 登録日 | 2006-08-08 | | 公開日 | 2006-09-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | 3,4,5-Trisubstituted Isoxazoles as Novel Ppardelta Agonists. Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1SZV
 
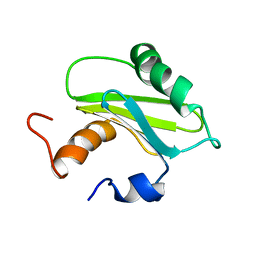 | | Structure of the Adaptor Protein p14 reveals a Profilin-like Fold with Novel Function | | 分子名称: | Late endosomal/lysosomal Mp1 interacting protein | | 著者 | Qian, C, Zhang, Q, Wang, X, Zeng, L, Farooq, A, Zhou, M.M. | | 登録日 | 2004-04-06 | | 公開日 | 2005-03-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Adaptor Protein p14 Reveals a Profilin-like Fold with Distinct Function
J.Mol.Biol., 347, 2005
|
|
7T47
 
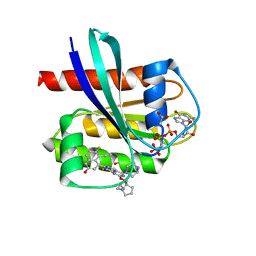 | | KRAS G12D (GppCp) with MRTX-1133 | | 分子名称: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)-5-ethynyl-6-fluoronaphthalen-2-ol, ACETATE ION, GLYCEROL, ... | | 著者 | Thomas, N.C, Gunn, R.J, Lawson, J.D, Wang, X, Matthew, M.A. | | 登録日 | 2021-12-09 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | A Non-covalent KRASG12D Allele Specific Inhibitor Demonstrates Potent Inhibition of KRAS-dependent Signaling and Regression of KRASG12D-mutant Tumors
Nature, 2022
|
|
7SOM
 
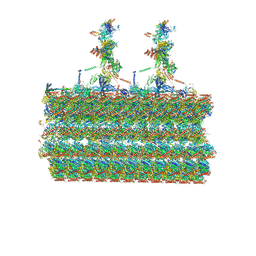 | | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | | 分子名称: | Cilia- and flagella-associated protein 20, FAP147, FAP178, ... | | 著者 | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | 登録日 | 2021-11-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8A2E
 
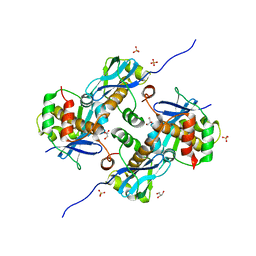 | | Crystal Structure of Human Parechovirus 3 2A protein | | 分子名称: | 2A protein, GLYCEROL, SULFATE ION | | 著者 | von Castelmur, E, Zhu, L, wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | 登録日 | 2022-06-03 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural plasticity of 2A proteins in the Parechovirus family.
To Be Published
|
|
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | 著者 | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | 登録日 | 2021-11-05 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8A2F
 
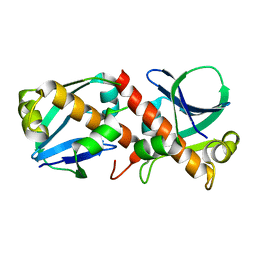 | | Crystal Structure of Ljunganvirus 1 2A protein | | 分子名称: | 2A protein | | 著者 | von Castelmur, E, Zhu, L, Wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | 登録日 | 2022-06-03 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structural plasticity of 2A proteins in the Parechovirus family
To Be Published
|
|
