1JWP
 
 | |
4QPI
 
 | | Crystal structure of hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
8IDI
 
 | | Crystal structure of nanobody VHH-T71 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T71, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
8IEE
 
 | |
8IDO
 
 | | Crystal structure of nanobody VHH-T148 with MERS-CoV RBD | | Descriptor: | Spike protein S1, VHH-T148, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
8IDM
 
 | | Crystal structure of nanobody VHH-227 with nanobody VHH-T71 and MERS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Definition of a Novel Nanobody Binding Site specifically targeting the MERS-CoV RBD Core-Domain with Neutralizing Capacity
To Be Published
|
|
8IFN
 
 | | MERS-CoV spike trimer in complex with nanobody VHH-T148 | | Descriptor: | Spike glycoprotein, VHH-T148, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X, Tian, L. | | Deposit date: | 2023-02-19 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and neutralizing mechanisms of camel nanobodies targeting the receptor-binding domain of MERS-CoV spike glycoprotein
To Be Published
|
|
6IZF
 
 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | (2R)-1-(dodecanoyloxy)-3-hydroxypropan-2-yl (5E,8E,11E)-tetradeca-5,8,11-trienoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, X.H, Zeng, Y, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IYZ
 
 | | Structural basis for activity of TRIC counter-ion channels in calcium release | | Descriptor: | CHLORIDE ION, Trimeric intracellular cation channel type A | | Authors: | Wang, X.H, Zeng, Y, Qin, L, Gao, F, Su, M, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8HXJ
 
 | | BANAL-20-52 Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Xu, G. | | Deposit date: | 2023-01-04 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The selective effect of fecal-oral transmission on the S proteins of bat SARS-CoV-2 related coronaviruses in favor of stability over infectivity
To Be Published
|
|
6IZ4
 
 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | Trimeric intracellular cation channel type B-B | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8HXK
 
 | | BANAL-20-236 S1 in complex with R. Affinis ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X, Xu, G. | | Deposit date: | 2023-01-04 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The selective effect of fecal-oral transmission on the S proteins of bat SARS-CoV-2 related coronaviruses in favor of stability over infectivity
To Be Published
|
|
7A1V
 
 | |
6XS6
 
 | | SARS-CoV-2 Spike D614G variant, minus RBD | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Egri, S.B, Dudkina, N, Luban, J, Shen, K. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.
Cell, 183, 2020
|
|
6NII
 
 | |
7P3O
 
 | |
7P3M
 
 | |
7P3P
 
 | |
1KRV
 
 | |
1KQA
 
 | |
6NJD
 
 | |
1KRU
 
 | |
1KRR
 
 | |
1AKN
 
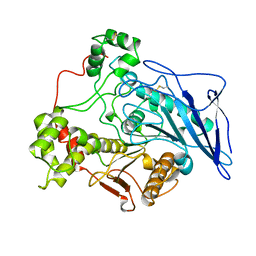 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-05-23 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
