5YIM
 
 | |
4O28
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-17 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1D
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.705 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4L4L
 
 | | Structural Analysis of a Phosphoribosylated Inhibitor in Complex with Human Nicotinamide Phosphoribosyltransferase | | 分子名称: | 1,2-ETHANEDIOL, 6-({4-[(3,5-difluorophenyl)sulfonyl]benzyl}carbamoyl)-1-(5-O-phosphono-beta-D-ribofuranosyl)imidazo[1,2-a]pyridin-1-ium, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Ho, Y, Zak, M, Liu, Y, Yuen, P, Zheng, X, Dragovich, S.P, Wang, W. | | 登録日 | 2013-06-08 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.122 Å) | | 主引用文献 | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
4JHR
 
 | | An auto-inhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs | | 分子名称: | G-protein-signaling modulator 2 | | 著者 | Pan, Z, Zhu, J, Shang, Y, Wei, Z, Jia, M, Xia, C, Wen, W, Wang, W, Zhang, M. | | 登録日 | 2013-03-05 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs
Structure, 21, 2013
|
|
4O10
 
 | | Structural and Biochemical Analyses of the Catalysis and Potency Impact of Inhibitor Phosphoribosylation by Human Nicotinamide Phosphoribosyltransferase | | 分子名称: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}indolizine-7-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Wang, W. | | 登録日 | 2013-12-14 | | 公開日 | 2014-06-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase.
Chembiochem, 15, 2014
|
|
6D5Y
 
 | |
6B16
 
 | | P21-activated kinase 1 in complex with a 4-azaindole inhibitor | | 分子名称: | N~4~-(5-cyclopropyl-1H-pyrazol-3-yl)-N~2~-[(1S)-1-(1H-pyrrolo[3,2-b]pyridin-5-yl)ethyl]pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PAK 1 | | 著者 | Rouge, L, Wang, W. | | 登録日 | 2017-09-16 | | 公開日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.285 Å) | | 主引用文献 | Synthesis and evaluation of a series of 4-azaindole-containing p21-activated kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5VFP
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFR
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFS
 
 | | Nucleotide-Driven Triple-State Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFU
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2018-07-18 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFT
 
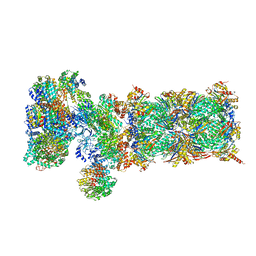 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFO
 
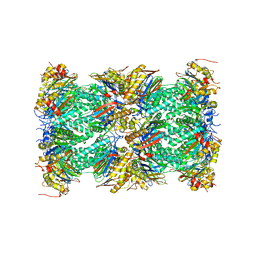 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-08 | | 公開日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VFQ
 
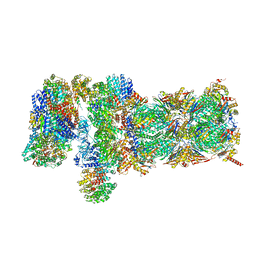 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
7YD2
 
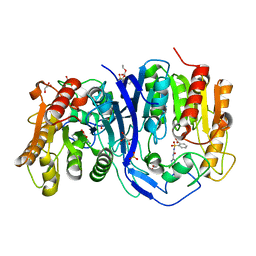 | | SulE_P44R_S209A | | 分子名称: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, ... | | 著者 | Liu, B, Ran, T, Wang, W, He, J. | | 登録日 | 2022-07-03 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7Y0L
 
 | | SulE-S209A | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | 著者 | Liu, B, Ran, T, He, J, Wang, W. | | 登録日 | 2022-06-05 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7VIB
 
 | | Crystal structure of human ACE2 and GX/P2V RBD | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | 著者 | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | 登録日 | 2021-09-26 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
8P5Z
 
 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | 分子名称: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | 著者 | Lau, K, Wang, W, Pojer, F, Larabi, A. | | 登録日 | 2023-05-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8P5Y
 
 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | 分子名称: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Lau, K, Wang, W, Pojer, F, Larabi, A. | | 登録日 | 2023-05-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7EW4
 
 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW2
 
 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | 分子名称: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW3
 
 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | 分子名称: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
4PPM
 
 | | Crystal structure of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14 | | 分子名称: | Aminotransferase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Lou, X.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | 登録日 | 2014-02-27 | | 公開日 | 2015-01-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the catalytic domain of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14
Biochem.Biophys.Res.Commun., 447, 2014
|
|
8JP3
 
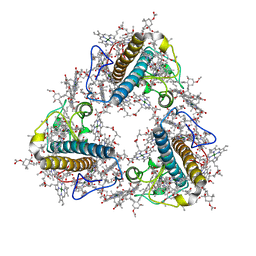 | | FCP trimer in diatom Thalassiosira pseudonana | | 分子名称: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | 著者 | Feng, Y, Li, Z, Zhou, C.C, Liu, C, Shen, J.R, Wang, W. | | 登録日 | 2023-06-10 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 2024
|
|
