3RAW
 
 | | Crystal Structure of human CDC-like kinase 3 isoform in complex with leucettine L41 | | Descriptor: | 5-(1,3-benzodioxol-5-ylmethyl)-2-(phenylamino)-4H-imidazol-4-one, Dual specificity protein kinase CLK3 | | Authors: | Filippakopoulos, P, Fedorov, O, King, O, Debdab, M, Carreaux, F, Renault, S, Bullock, A, Muniz, J.R.C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Meijer, L, Bazureau, J.P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of human CDC-like kinase 3 isoform with a benzo-dioxol ligand
To be Published
|
|
3QF9
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand | | Descriptor: | 6-{5-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]furan-2-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}naphthalene-1-carboxamide, CHLORIDE ION, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Miduturu, C.V, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand
To be Published
|
|
3GP0
 
 | | Crystal Structure of Human Mitogen Activated Protein Kinase 11 (p38 beta) in complex with Nilotinib | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 11, ... | | Authors: | Filippakopoulos, P, Barr, A, Fedorov, O, Keates, T, Soundararajan, M, Elkins, J, Salah, E, Burgess-Brown, N, Ugochukwu, E, Pike, A.C.W, Muniz, J, Roos, A, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Mitogen Activated Protein Kinase 11 (p38 beta) in complex with Nilotinib
To be Published
|
|
2OS2
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys36 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OT7
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide monomethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-07 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
5F5C
 
 | | Crystal Structure of human JMJD2D complexed with KDOPP7 | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(phenylmethyl)amino]methyl]-1~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3RMU
 
 | | Crystal structure of human Methylmalonyl-CoA epimerase, MCEE | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Methylmalonyl-CoA epimerase, ... | | Authors: | Chaikuad, A, Krysztofinska, E, Froese, D.S, Yue, W.W, Vollmar, M, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Methylmalonyl-CoA epimerase, MCEE
To be Published
|
|
5AMF
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment Ethyl 4,5,6,7- tetrahydro-1H-indazole-5-carboxylate (SGC - Diamond I04-1 fragment screening) | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 1, ETHYL (5R)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE-5-CARBOXYLATE, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment Ethyl 4,5,6,7-Tetrahydro-1H-Indazole-5-Carboxylate
To be Published
|
|
5AME
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment 4-acetyl- piperazin-2-one (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 4-acetyl-piperazin-2-one, BROMODOMAIN-CONTAINING PROTEIN 1, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment 4-Acetyl-Piperazin-2-One
To be Published
|
|
5F5A
 
 | | Crystal Structure of human JMJD2D complexed with KDOAM16 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(furan-2-ylmethylamino)methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4D, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Bradley, A.R, Szykowska, A, Ruda, G.F, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3RMW
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) T83M mutant complexed with manganese and UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MAGNESIUM ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Yue, W.W, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5F5I
 
 | | Crystal Structure of human JMJD2A complexed with KDOOA011340 | | Descriptor: | 2-[[(phenylmethyl)amino]methyl]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Krojer, T, Vollmar, M, Crawley, L, Szykowska, A, Gileadi, C, Johansson, C, England, K, Yang, H, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2OQ6
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
3OOM
 
 | | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507 | | Descriptor: | 1,2-ETHANEDIOL, 1-{3-[6-(tetrahydro-2H-pyran-4-ylamino)imidazo[1,2-b]pyridazin-3-yl]phenyl}ethanone, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ACVR1 kinase domain in complex with the imidazo[1,2-b]pyridazine inhibitor K00507
To be Published
|
|
3OOY
 
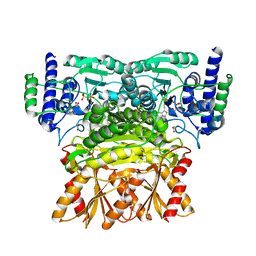 | | Crystal structure of human Transketolase (TKT) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Krojer, T, Krysztofinska, E, Guo, K, Pilka, E, Kochan, G, Chaikuad, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Transketolase (TKT)
To be Published
|
|
2OQ7
 
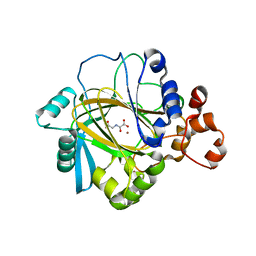 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
3P1F
 
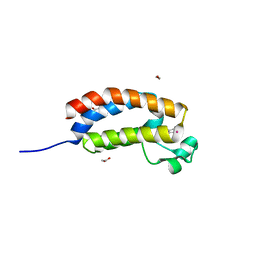 | | Crystal structure of the bromodomain of human CREBBP in complex with a hydroquinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, CREB-binding protein, ... | | Authors: | Filippakopoulos, P, Picaud, S, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with a hydroquinazolin ligand
To be Published
|
|
3S95
 
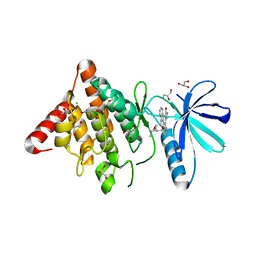 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
3QNF
 
 | | Crystal structure of the open state of human endoplasmic reticulum aminopeptidase 1 ERAP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, ZINC ION, ... | | Authors: | Vollmar, M, Kochan, G, Krojer, T, Harvey, D, Chaikuad, A, Allerston, C, Muniz, J.R.C, Raynor, J, Ugochukwu, E, Berridge, G, Wordsworth, B.P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2OX0
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide dimethylated at Lys9 | | Descriptor: | CHLORIDE ION, JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
3S91
 
 | | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain-containing protein 3, ISOPROPYL ALCOHOL | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1
To be Published
|
|
2PD6
 
 | | Structure of human hydroxysteroid dehydrogenase type 8, HSD17B8 | | Descriptor: | Estradiol 17-beta-dehydrogenase 8, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Turnbull, A.P, Salah, E, Gileadi, O, Savitsky, P, Guo, K, Bunkoczi, G, Pike, A.C.W, Ugochukwu, E, Umeano, C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-31 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human hydroxysteroid dehydrogenase type 8, HSD17B8
To be Published
|
|
2PLA
 
 | | Crystal structure of human glycerol-3-phosphate dehydrogenase 1-like protein | | Descriptor: | CHLORIDE ION, Glycerol-3-phosphate dehydrogenase 1-like protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Uppenberg, J, Smee, C, Hozjan, V, Kavanagh, K, Bunkoczi, G, Papagrigoriou, E, Pike, A.C.W, Ugochukwu, E, Umeano, C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of human glycerol-3-phosphate dehydrogenase 1-like protein.
To be Published
|
|
2Q0N
 
 | | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Turnbull, A, Papagrigoriou, E, Pike, A.W, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide.
To be Published
|
|
2PN8
 
 | | Crystal structure of human peroxiredoxin 4 (thioredoxin peroxidase) | | Descriptor: | Peroxiredoxin-4 | | Authors: | Pilka, E.S, Kavanagh, K.L, Cooper, C, von Delft, F, Sundstrom, M, Arrowsmith, C.A, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-24 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human peroxiredoxin 4 (thioredoxin peroxidase).
To be Published
|
|
