1IIB
 
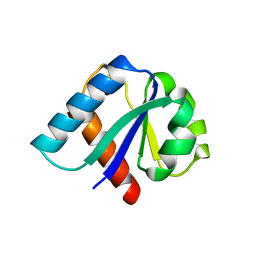 | | CRYSTAL STRUCTURE OF IIBCELLOBIOSE FROM ESCHERICHIA COLI | | Descriptor: | ENZYME IIB OF THE CELLOBIOSE-SPECIFIC PHOSPHOTRANSFERASE SYSTEM | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Reizer, J, Saier, M.H, Thunnissen, M.M.G.M, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1996-12-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an energy-coupling protein from bacteria, IIBcellobiose, reveals similarity to eukaryotic protein tyrosine phosphatases.
Structure, 5, 1997
|
|
1A3A
 
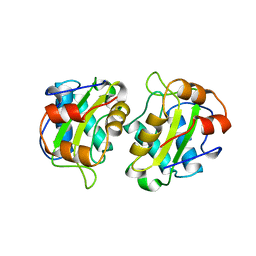 | | CRYSTAL STRUCTURE OF IIA MANNITOL FROM ESCHERICHIA COLI | | Descriptor: | MANNITOL-SPECIFIC EII | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Hangyi, I, Kouwijzer, M.L.C.E, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1998-01-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the Escherichia coli phosphotransferase IIAmannitol reveals a novel fold with two conformations of the active site.
Structure, 6, 1998
|
|
1OEV
 
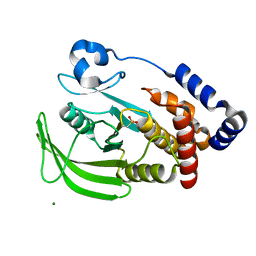 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
1OEU
 
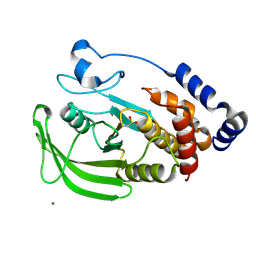 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
1OET
 
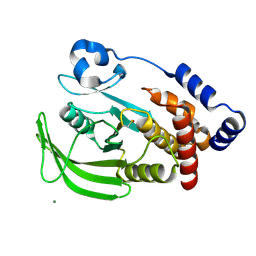 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | -TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1, MAGNESIUM ION | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
1OES
 
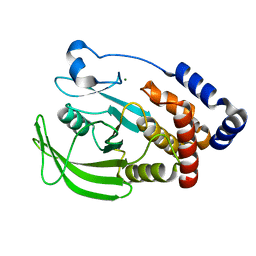 | | Oxidation state of protein tyrosine phosphatase 1B | | Descriptor: | MAGNESIUM ION, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | van Montfort, R.L.M, Congreve, M, Tisi, D, Carr, R, Jhoti, H. | | Deposit date: | 2003-03-31 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B.
Nature, 423, 2003
|
|
1OKI
 
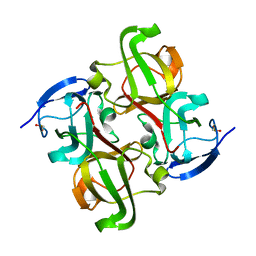 | |
1GME
 
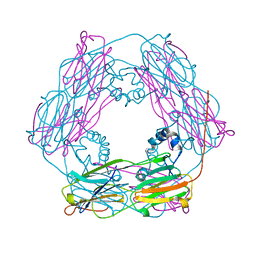 | | Crystal structure and assembly of an eukaryotic small heat shock protein | | Descriptor: | HEAT SHOCK PROTEIN 16.9B | | Authors: | Van Montfort, R.L.M, Basha, E, Friedrich, K.L, Slingsby, C, Vierling, E. | | Deposit date: | 2001-09-13 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Assembly of an Eukaryotic Small Heat Shock Protein
Nat.Struct.Biol., 8, 2001
|
|
8OIZ
 
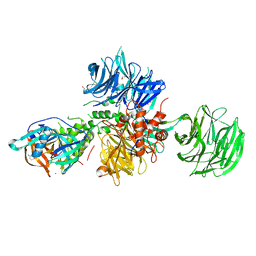 | | Crystal structure of human CRBN-DDB1 in complex with Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Le Bihan, Y.-V, Cabry, M.P, van Montfort, R.L.M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
8OJH
 
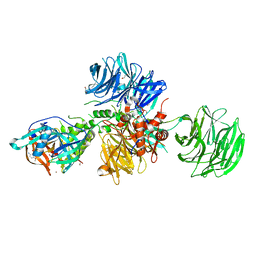 | | Crystal structure of human CRBN-DDB1 in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-7-methoxy-isoindole-1,3-dione, DNA damage-binding protein 1, ... | | Authors: | Cabry, M.P, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
8C78
 
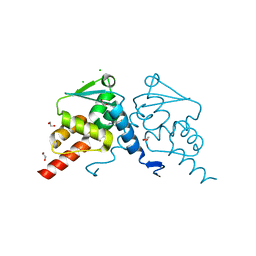 | | Crystal structure of human BCL6 BTB domain in complex with compound CCT374705 | | Descriptor: | (2~{S})-10-[(3-chloranyl-2-fluoranyl-pyridin-4-yl)amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an In Vivo Chemical Probe for BCL6 Inhibition by Optimization of Tricyclic Quinolinones.
J.Med.Chem., 66, 2023
|
|
6H4P
 
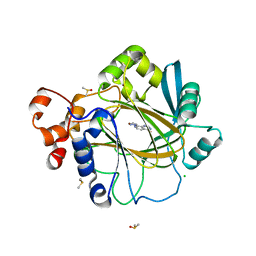 | | Crystal structure of human KDM4A in complex with compound 16a | | Descriptor: | 8-[4-[2-[4-(3-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4Y
 
 | | Crystal structure of human KDM4A in complex with compound 17e | | Descriptor: | 8-[4-[2-[4-[4-(2-morpholin-4-ylethyl)phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
7Q4R
 
 | | Crystal structure of human HSP72-NBD in complex with fragment 1 | | Descriptor: | 4-(4-phenyl-1,3-thiazol-2-yl)piperazine-1-carboxamide, Heat shock 70 kDa protein 1A, MAGNESIUM ION, ... | | Authors: | Le Bihan, Y.V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and Characterization of a Cryptic Secondary Binding Site in the Molecular Chaperone HSP70.
Molecules, 27, 2022
|
|
5MKS
 
 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in down-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MKR
 
 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in up-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, CITRATE ANION, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4BDF
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 5-METHYL-3-PHENYL-1H-PYRAZOLE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
7Q7S
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-(ethylamino)-1,3-dimethyl-2-oxidanylidene-quinolin-6-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7Q7V
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 12a | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[(2R)-2-cyclopropyl-7-methyl-6-oxidanylidene-1,2,3,4-tetrahydro-[1,4]oxazepino[2,3-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7Q7U
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 9a | | Descriptor: | 2-chloranyl-4-[[(2S)-2,7-dimethyl-6-oxidanylidene-1,2,3,4-tetrahydro-[1,4]oxazepino[2,3-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7Q7T
 
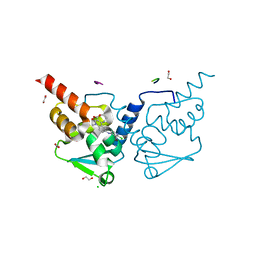 | | Crystal structure of human BCL6 BTB domain in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[(2S)-2,7-dimethyl-5,6-bis(oxidanylidene)-2,3-dihydro-1H-[1,4]oxazepino[6,5-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7QK0
 
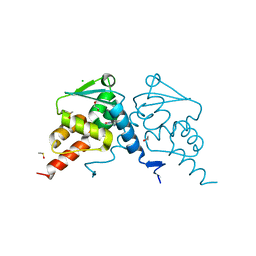 | | Crystal structure of human BCL6 BTB domain in complex with compound 12a | | Descriptor: | (2~{S})-10-[[5-chloranyl-2-[(3~{S},5~{R})-3-methyl-5-oxidanyl-piperidin-1-yl]pyrimidin-4-yl]amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Improved Binding Affinity and Pharmacokinetics Enable Sustained Degradation of BCL6 In Vivo .
J.Med.Chem., 65, 2022
|
|
7Q7R
 
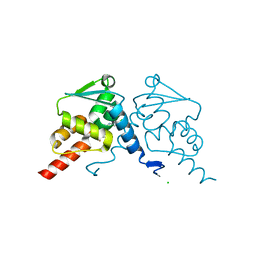 | | Crystal structure of human BCL6 BTB domain in complex with compound 1 | | Descriptor: | 2-chloranyl-4-[[(2S)-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-6-oxidanylidene-2,4-dihydro-1H-[1,4]oxazepino[2,3-c]quinolin-10-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein, CHLORIDE ION | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimizing Shape Complementarity Enables the Discovery of Potent Tricyclic BCL6 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZWV
 
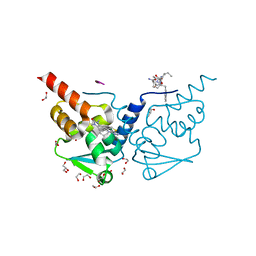 | | Crystal structure of human BCL6 BTB domain in complex with compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-5-[[6-(phenylmethylsulfanyl)pyrimidin-4-yl]amino]benzimidazol-2-one, ALA-TRP-VAL-ILE-PRO-ALA, ... | | Authors: | Collie, G.W, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
7ZWQ
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 10 | | Descriptor: | 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays.
Sci Rep, 12, 2022
|
|
