3UM9
 
 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Bpro0530 | | Descriptor: | Haloacid dehalogenase, type II, NICKEL (II) ION, ... | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
4O2I
 
 | | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium | | Descriptor: | Non-LEE encoded type III effector C, ZINC ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium
To be Published
|
|
5HNM
 
 | | Crystal structure of vancomycin resistance D,D-pentapeptidase VanY E175A mutant from VanB-type resistance cassette in complex with Zn(II) | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, SULFATE ION, ZINC ION | | Authors: | Stogios, P.J, Chun, J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
6PI9
 
 | | Crystal structure of 16S rRNA methyltransferase RmtF in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Stogios, P.J, Kim, Y, Evdokimova, E, Di Leo, R, Semper, C, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 16S rRNA methylase RmtF in complex with S-Adenosyl-L-homocysteine
To be Published
|
|
3S40
 
 | | The crystal structure of a diacylglycerol kinases from Bacillus anthracis str. Sterne | | Descriptor: | diacylglycerol kinase | | Authors: | Tan, K, Zhang, R, Xu, X, Cui, H, Peterson, S, Savchenko, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a diacylglycerol kinases from Bacillus anthracis str. Sterne
To be Published
|
|
5JD5
 
 | | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy | | Descriptor: | CHLORIDE ION, MGS-MilE3 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Martinez-Martinez, M, Chernikova, T.N, Golyshin, P.N, Yakimov, M.M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy
To Be Published
|
|
3UMC
 
 | | Crystal Structure of the L-2-Haloacid Dehalogenase PA0810 | | Descriptor: | CHLORIDE ION, SODIUM ION, haloacid dehalogenase | | Authors: | Petit, P, Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-13 | | Release date: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UMG
 
 | | Crystal Structure of the Defluorinating L-2-Haloacid Dehalogenase Rha0230 | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase | | Authors: | Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-13 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UZR
 
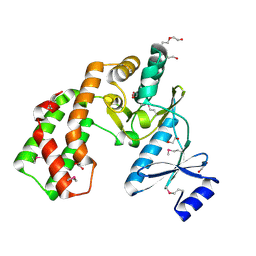 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside phosphotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form
TO BE PUBLISHED
|
|
4NE4
 
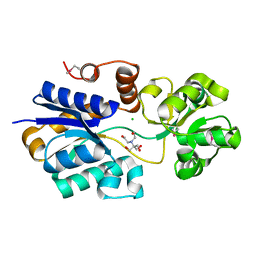 | | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, substrate binding protein (Proline/glycine/betaine), ... | | Authors: | Tkaczuk, K.L, Nicholls, R, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB
To be Published
|
|
3T32
 
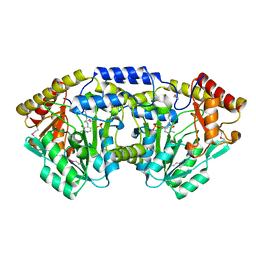 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
3UWD
 
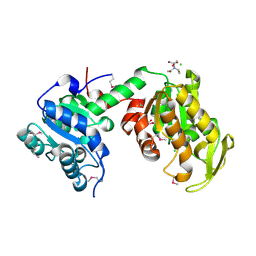 | | Crystal Structure of Phosphoglycerate Kinase from Bacillus Anthracis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zheng, H, Chruszcz, M, Porebski, P, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-01 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
4QL5
 
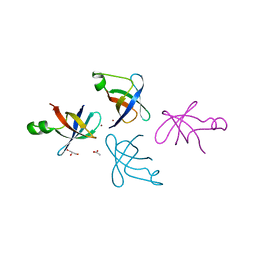 | | Crystal structure of translation initiation factor IF-1 from Streptococcus pneumoniae TIGR4 | | Descriptor: | ACETATE ION, GLYCEROL, Translation initiation factor IF-1, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Crystal structure of translation initiation factor IF-1 from Streptococcus pneumoniae TIGR4
TO BE PUBLISHED
|
|
3TTG
 
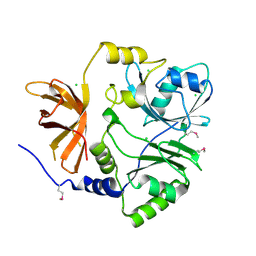 | | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, Putative aminomethyltransferase | | Authors: | Michalska, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum
TO BE PUBLISHED
|
|
3TYK
 
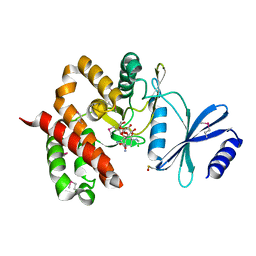 | | Crystal structure of aminoglycoside phosphotransferase APH(4)-Ia | | Descriptor: | CHLORIDE ION, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | Authors: | Stogios, P.J, Shabalin, I.G, Shakya, T, Evdokmova, E, Fan, Y, Chruszcz, M, Minor, W, Wright, G.D, Savchenko, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme.
J.Biol.Chem., 286, 2011
|
|
4M3S
 
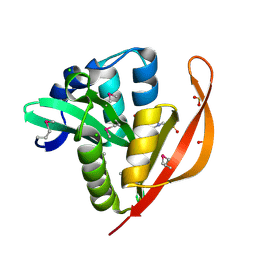 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Xu, X, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-08-06 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Double trouble-Buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments.
Protein Sci., 23, 2014
|
|
1Z6N
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA1234 from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, hypothetical protein PA1234 | | Authors: | Zhang, R, Xu, L, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1234 from Pseudomonas aeruginosa
To be Published
|
|
1ZL0
 
 | | Structure of Protein of Unknown Function PA5198 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-04 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA5198 at 1.1 A resolution.
To be Published
|
|
1ZKI
 
 | | Structure of conserved protein PA5202 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, hypothetical protein PA5202 | | Authors: | Cuff, M.E, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
3TYR
 
 | | Crystal structure of transcriptional regulator VanUg, Form I | | Descriptor: | Transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Dong, A, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form I
TO BE PUBLISHED
|
|
4WH5
 
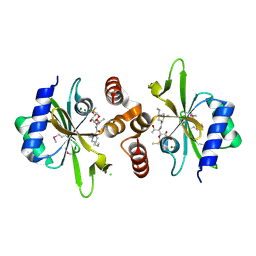 | | Crystal structure of lincosamide antibiotic adenylyltransferase LnuA, lincomycin-bound | | Descriptor: | CHLORIDE ION, LINCOMYCIN, Lincosamide resistance protein, ... | | Authors: | Stogios, P.J, Dong, A, Minasov, G, Evdokimova, E, Egorova, O, Kudritska, M, Yim, O, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-20 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | CRYSTAL STRUCTURE OF LINCOSAMIDE ANTIBIOTIC ADENYLYLTRANSFERASE LNUA, LINCOMYCIN BOUND
To Be Published
|
|
2LOJ
 
 | | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273 | | Descriptor: | Putative cytoplasmic protein | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of TSTM1273 from Salmonella typhimurium LT2, NESG target STT322, CSGID target IDP01027 and OCSP target TSTM1273
To be Published
|
|
3T4X
 
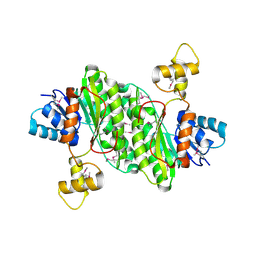 | | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Oxidoreductase, short chain dehydrogenase/reductase family | | Authors: | Filippova, E.V, Wawrzak, Z, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-17 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
2M46
 
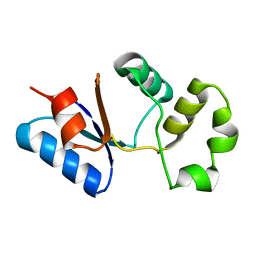 | | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841 | | Descriptor: | Arsenate reductase, putative | | Authors: | Wu, B, Yee, A, Houliston, S, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SACOL0876 from Staphylococcus aureus COL, NESG target ZR353 and CSGID target IDP00841
To be Published
|
|
3SKS
 
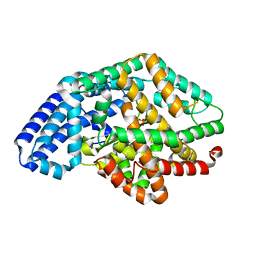 | | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames | | Descriptor: | PHOSPHATE ION, Putative Oligoendopeptidase F, ZINC ION | | Authors: | Wajerowicz, W, Onopriyenko, O, Porebski, P, Domagalski, M, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
