4KFV
 
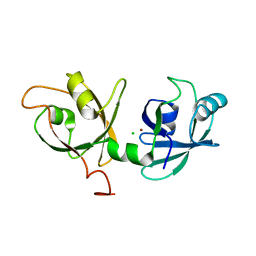 | |
3TGV
 
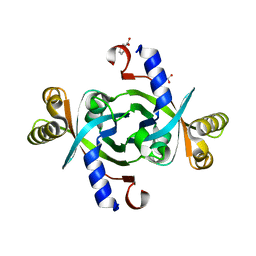 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
5GQQ
 
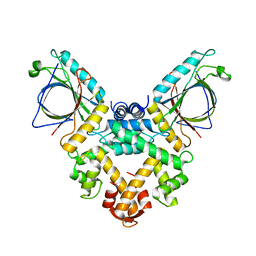 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
4NXQ
 
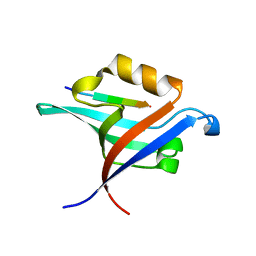 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Caspr4 Peptide | | Descriptor: | Contactin-associated protein-like 4 peptide, T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
4NXP
 
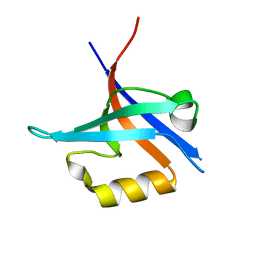 | | Crystal Structure of Free T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
4NXR
 
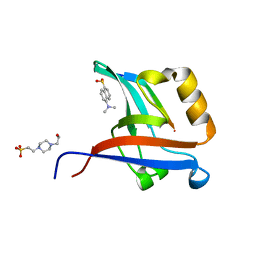 | | Crystal Structure of T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) in Complex With Neurexin-1 Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(DIMETHYLAMINO)-1-NAPHTHALENESULFONIC ACID(DANSYL ACID), Neurexin-2-beta Peptide, ... | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
6AAX
 
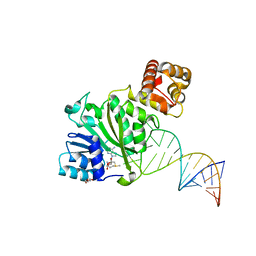 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AJK
 
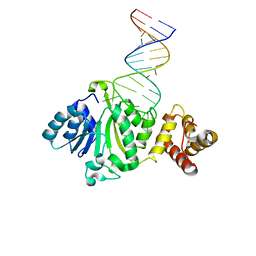 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
4KC3
 
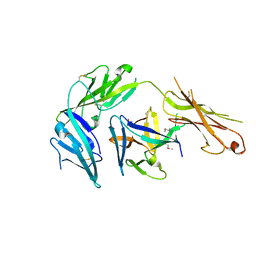 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8GUB
 
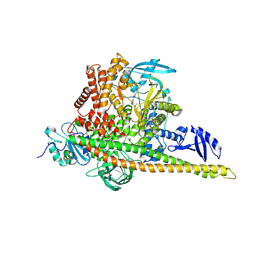 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUD
 
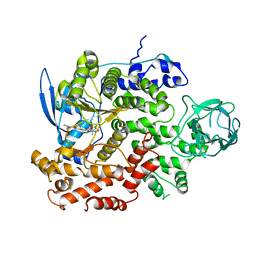 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUA
 
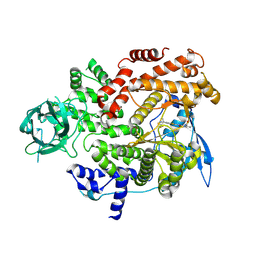 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E542K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6JYW
 
 | |
4HBL
 
 | | Crystal structure of AbfR of Staphylococcus epidermidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Liu, X, Sun, X, Gan, J, Lan, L, Yang, C.-G. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-02 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation-sensing Regulator AbfR Regulates Oxidative Stress Responses, Bacterial Aggregation, and Biofilm Formation in Staphylococcus epidermidis.
J.Biol.Chem., 288, 2013
|
|
5Y0I
 
 | | Solution structure of arenicin-3 derivative N1 | | Descriptor: | NZ17074(N1) | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5Y0H
 
 | | Solution structure of arenicin-3 derivative N6 | | Descriptor: | N6 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
5Y0J
 
 | | Solution structure of arenicin-3 derivative N2 | | Descriptor: | N2 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
3S17
 
 | | RNA Polymerase II Initiation Complex with a 9-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S15
 
 | | RNA Polymerase II Initiation Complex with a 7-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S2H
 
 | | RNA Polymerase II Initiation Complex with a 6-nt RNA containing a 2[prime]-iodo ATP | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S1M
 
 | | RNA Polymerase II Initiation Complex with a 5-nt RNA (variant 1) | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*GP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-15 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
4QBA
 
 | |
6JC0
 
 | |
7CYF
 
 | | Cryo-EM structure of bicarbonate transporter SbtA in complex with PII-like signaling protein SbtB from Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION, ... | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
7CYE
 
 | | Cryo-EM structure of sodium-dependent bicarbonate transporter SbtA from Synechocystis sp. PCC 6803 | | Descriptor: | Slr1512 protein | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
