7S72
 
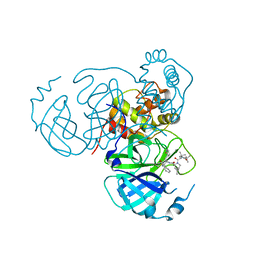 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI36 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S75
 
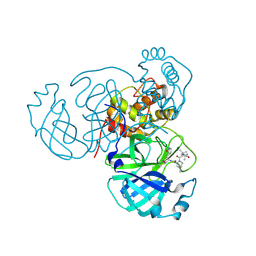 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI42 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6W
 
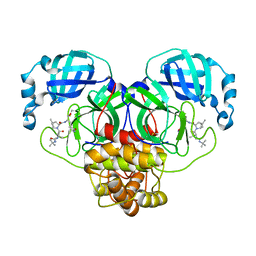 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
4CKV
 
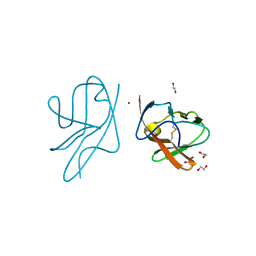 | | Crystal structure of VEGFR-1 domain 2 in presence of Zn | | Descriptor: | 1,2-ETHANEDIOL, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1, ZINC ION | | Authors: | Gaucher, J.-F, Reille-Seroussi, M, Gagey-Eilstein, N, Broussy, S, Coric, P, Seijo, B, Lascombe, M.-B, Gautier, B, Liu, W.-Q, Huguenot, F, Inguimbert, N, Bouaziz, S, Vidal, M, Broutin, I. | | Deposit date: | 2014-01-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Biophysical Studies of the Induced Dimerization of Human Vegf R Receptor 1 Binding Domain by Divalent Metals Competing with Vegf-A
Plos One, 11, 2016
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
7KX0
 
 | | Crystal structure of the CD27:CD70 co-stimulatory complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD27 antigen, ... | | Authors: | Maben, Z, Liu, W, Mosyak, L, Chaparro-Riggers, J. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural delineation and phase-dependent activation of the costimulatory CD27:CD70 complex.
J.Biol.Chem., 297, 2021
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
5UVJ
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
3S8G
 
 | | 1.8 A structure of ba3 cytochrome c oxidase mutant (A120F) from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
3S8F
 
 | | 1.8 A structure of ba3 cytochrome c oxidase from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
7S6Y
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI32 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S74
 
 | |
7S6Z
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7LMC
 
 | |
7S73
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI37 | | Descriptor: | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name), 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S70
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI34 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(butylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7EL7
 
 | | NMR solution structure of the 1:1 complex of a quadruplex-duplex hybrid MYT1L and a platinum(II) ligand L1Pt(dien) | | Descriptor: | G-quadruplex DNA MYT1L, Pt(diethylenetriamine)(2-(pyridin-4-ylmethyl)benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Liu, W, Mao, Z.-W. | | Deposit date: | 2021-04-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spatial Matching Selectivity and Solution Structure of Organic-Metal Hybrid to Quadruplex-Duplex Hybrid.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7TLL
 
 | | Structure of SARS-CoV-2 Mpro Omicron P132H in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-01-26 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
5F67
 
 | | An exquisitely specific PDZ/target recognition revealed by the structure of INAD PDZ3 in complex with TRP channel tail | | Descriptor: | Inactivation-no-after-potential D protein, TRP C terminal Tail | | Authors: | Ye, F, Shang, Y, Liu, W, Zhang, M. | | Deposit date: | 2015-12-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An Exquisitely Specific PDZ/Target Recognition Revealed by the Structure of INAD PDZ3 in Complex with TRP Channel Tail
Structure, 24, 2016
|
|
6IJ1
 
 | | Crystal structure of a protein from Actinoplanes | | Descriptor: | ACETATE ION, IMIDAZOLE, Prenylcyclase | | Authors: | Yang, Z.Z, Zhang, L.L, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-10-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Crystal structure of TchmY from Actinoplanes teichomyceticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5HDG
 
 | |
5HDN
 
 | |
4TKN
 
 | | Structure of the SNX17 FERM domain bound to the second NPxF motif of KRIT1 | | Descriptor: | Krev interaction trapped protein 1, Sorting nexin-17 | | Authors: | Stiegler, A.L, Zhang, R, Liu, W, Boggon, T.J. | | Deposit date: | 2014-05-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Determinants for Binding of Sorting Nexin 17 (SNX17) to the Cytoplasmic Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 289, 2014
|
|
8V4U
 
 | |
6J6W
 
 | | Mutant K23N of heat shock factor 4-DBD | | Descriptor: | Heat shock factor protein 4, NITRATE ION, SODIUM ION | | Authors: | Xiao, Z.Y, Cui, L.W, Wang, S, Liu, W. | | Deposit date: | 2019-01-16 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structure of the mutant K23N of heat shock factor 4-DBD at 1.69 Angstroms resolution.
To Be Published
|
|
