4OCA
 
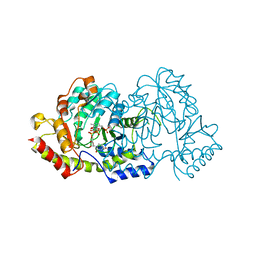 | | Cryatal structure of ArnB K188A complexted with PLP and UDP-Ara4N | | 分子名称: | (2R,3R,4S,5S)-3,4-dihydroxy-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]tetrahydro-2H-pyr an-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate aminotransferase | | 著者 | Sousa, M.C, Lee, M. | | 登録日 | 2014-01-08 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for Substrate Specificity in ArnB. A Key Enzyme in the Polymyxin Resistance Pathway of Gram-Negative Bacteria.
Biochemistry, 53, 2014
|
|
3LZD
 
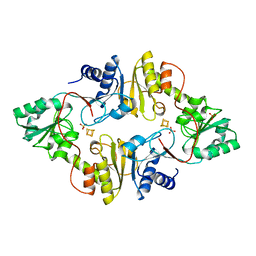 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | 分子名称: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | 著者 | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | 登録日 | 2010-03-01 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
3TVQ
 
 | | Crystal structure of TCM Aro/Cyc complexed with trans-dihidroquercetin | | 分子名称: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, Multifunctional cyclase-dehydratase-3-O-methyl transferase tcmN | | 著者 | Ames, B.D, Lee, M.-Y, Tsai, S.-C. | | 登録日 | 2011-09-20 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
3NIL
 
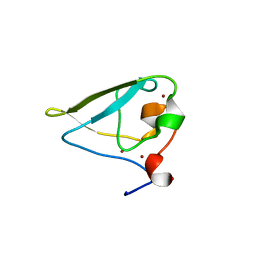 | | The structure of UBR box (RDAA) | | 分子名称: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
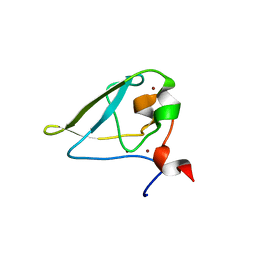 | | The structure of UBR box (REAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | 分子名称: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIM
 
 | | The structure of UBR box (RRAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
5FDN
 
 | |
5Y2T
 
 | | Structure of PPARgamma ligand binding domain - lobeglitazone complex | | 分子名称: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | 著者 | Im, Y.J, Lee, M. | | 登録日 | 2017-07-27 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
5Y2O
 
 | | Structure of PPARgamma ligand binding domain-pioglitazone complex | | 分子名称: | (5S)-5-[[4-[2-(5-ethylpyridin-2-yl)ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | 著者 | Im, Y.J, Lee, M. | | 登録日 | 2017-07-26 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
3NIJ
 
 | | The structure of UBR box (HIAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
 | | The structure of UBR box (RLGES) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
 | | The structure of UBR box (native1) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EPN
 
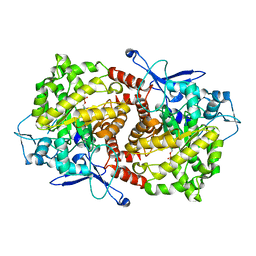 | | Crystal structure of Caulobacter crescentus ThiC complexed with imidazole ribonucleotide | | 分子名称: | 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, Thiamine biosynthesis protein thiC | | 著者 | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | 登録日 | 2008-09-29 | | 公開日 | 2008-10-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
3NIH
 
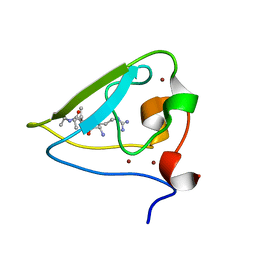 | | The structure of UBR box (RIAAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
1M6F
 
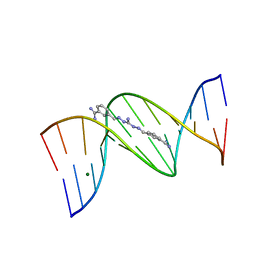 | | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove | | 分子名称: | 3-[C-[N'-(3-CARBAMIMIDOYL-BENZYLIDENIUM)-HYDRAZINO]-[[AMINOMETHYLIDENE]AMINIUM]-IMINOMETHYL]-BENZAMIDINIUM, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Nguyen, B, Lee, M.P.H, Hamelberg, D, Joubert, A, Bailly, C, Brun, R, Neidle, S, Wilson, W.D. | | 登録日 | 2002-07-16 | | 公開日 | 2002-11-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove
J.Am.Chem.Soc., 124, 2002
|
|
3NII
 
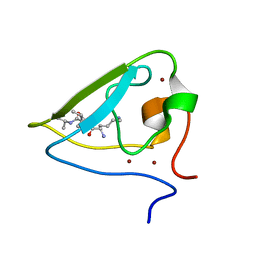 | | The structure of UBR box (KIAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EPO
 
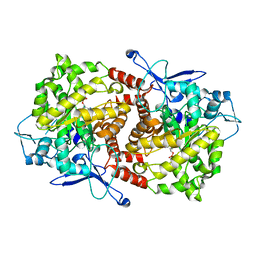 | | Crystal structure of Caulobacter crescentus ThiC complexed with HMP-P | | 分子名称: | (4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL DIHYDROGEN PHOSPHATE, Thiamine biosynthesis protein thiC | | 著者 | Li, S, Chatterjee, A, Zhang, Y, Grove, T.L, Lee, M, Krebs, C, Booker, S.J, Begley, T.P, Ealick, S.E. | | 登録日 | 2008-09-29 | | 公開日 | 2008-10-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily
Nat.Chem.Biol., 4, 2008
|
|
7EAR
 
 | |
1XOY
 
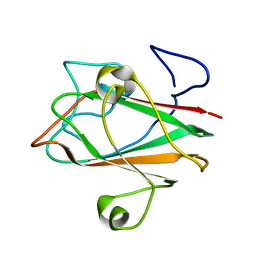 | | Solution structure of At3g04780.1, an Arabidopsis ortholog of the C-terminal domain of human thioredoxin-like protein | | 分子名称: | hypothetical protein At3g04780.1 | | 著者 | Song, J, Robert, C.T, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2004-10-07 | | 公開日 | 2004-10-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of At3g04780.1-des15, an Arabidopsis thaliana ortholog of the C-terminal domain of human thioredoxin-like protein.
Protein Sci., 14, 2005
|
|
1VZK
 
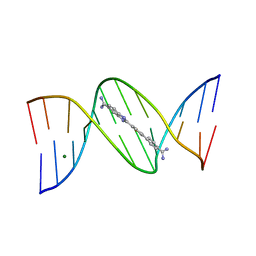 | | A Thiophene Based Diamidine Forms a "Super" AT Binding Minor Groove Agent | | 分子名称: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-THIENYL)-1H-BENZIMIDAZOLE-6- CARBOXIMIDAMIDE DIHYDROCHLORIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP *CP*G)-3', MAGNESIUM ION | | 著者 | Mallena, S, Lee, M.P.H, Bailly, C, Neidle, S, Kumar, A, Boykin, D.W, Wilson, W.D. | | 登録日 | 2004-05-20 | | 公開日 | 2004-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Thiophene-Based Diamidine Forms a "Super" at Binding Minor Groove Agent
J.Am.Chem.Soc., 142, 2004
|
|
1YDU
 
 | | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain | | 分子名称: | At5g01610 | | 著者 | Zhao, Q, Cornilescu, C.C, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2004-12-26 | | 公開日 | 2005-02-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain
To be Published
|
|
3SEP
 
 | | E. coli (lacZ) beta-galactosidase (S796A) | | 分子名称: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | 登録日 | 2011-06-10 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
1YYC
 
 | |
3T0B
 
 | | E. coli (LacZ) beta-galactosidase (S796T) IPTG complex | | 分子名称: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | 著者 | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | 登録日 | 2011-07-19 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | er-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
