9ERK
 
 | |
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
9ERI
 
 | |
9ERJ
 
 | |
9ERL
 
 | |
8BEW
 
 | |
7M6R
 
 | |
7M6S
 
 | |
7M6O
 
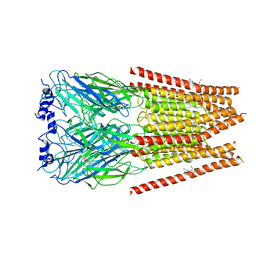 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7C9B
 
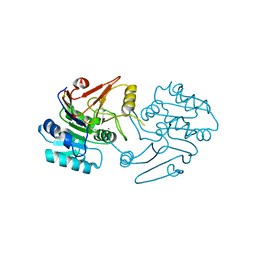 | | Crystal structure of dipeptidase-E from Xenopus laevis | | Descriptor: | Alpha-aspartyl dipeptidase, CALCIUM ION, SODIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
8JCS
 
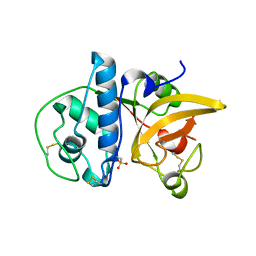 | |
8JCR
 
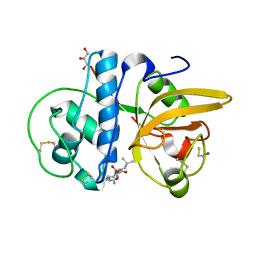 | | Crystal structure of Calotropain FI from Calotropis gigantea (pH 6.0) | | Descriptor: | GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, Procerain, ... | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Papain-Like Cysteine Protease, Calotropain FI, Purified from the Latex of Calotropis gigantea.
J.Agric.Food Chem., 73, 2025
|
|
8JCQ
 
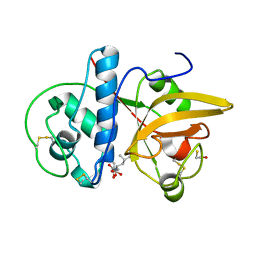 | | Crystal structure of calotropain FI from Calotropis gigantea | | Descriptor: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, Procerain | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Papain-Like Cysteine Protease, Calotropain FI, Purified from the Latex of Calotropis gigantea.
J.Agric.Food Chem., 73, 2025
|
|
7FFP
 
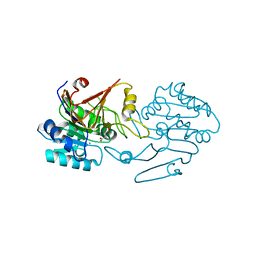 | | Crystal structure of di-peptidase-E from Xenopus laevis | | Descriptor: | ASPARTIC ACID, Alpha-aspartyl dipeptidase, CALCIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
6UD3
 
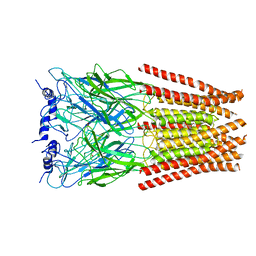 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/PTX-bound open/blocked conformation | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Basak, S, Chakrapani, S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
5Y05
 
 | |
7VRR
 
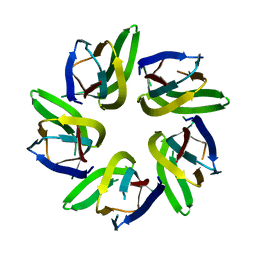 | |
5ID2
 
 | |
8QZD
 
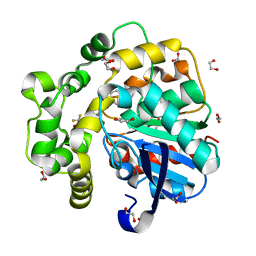 | | Soluble epoxide hydrolase in complex with Epoxykinin | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-bromanyl-3-[2,2,2-tris(fluoranyl)ethanoyl]indol-1-yl]-N-cycloheptyl-ethanamide, BROMIDE ION, ... | | Authors: | Kumar, A, Ehrler, J.M.H, Ziegler, S, Doetsch, L, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the sEH Inhibitor Epoxykynin as a Potent Kynurenine Pathway Modulator.
J.Med.Chem., 67, 2024
|
|
4K55
 
 | |
7NPW
 
 | |
1JYM
 
 | | Crystals of Peptide Deformylase from Plasmodium falciparum with Ten Subunits per Asymmetric Unit Reveal Critical Characteristics of the Active Site for Drug Design | | Descriptor: | COBALT (II) ION, Peptide Deformylase | | Authors: | Kumar, A, Nguyen, K.T, Srivathsan, S, Ornstein, B, Turley, S, Hirsh, I, Pei, D, Hol, W.G.J. | | Deposit date: | 2001-09-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystals of peptide deformylase from Plasmodium falciparum reveal critical characteristics of the active site for drug design.
Structure, 10, 2002
|
|
7CXC
 
 | |
7CXD
 
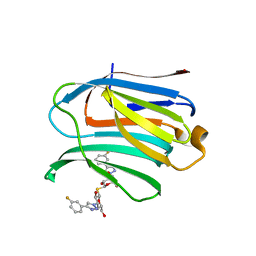 | | Xray structure of rat Galectin-3 CRD in complex with TD-139 belonging to P121 space group | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, BROMIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXB
 
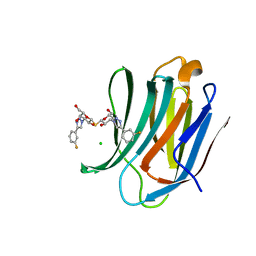 | | Structure of mouse Galectin-3 CRD in complex with TD-139 belonging to P6522 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
