5ILW
 
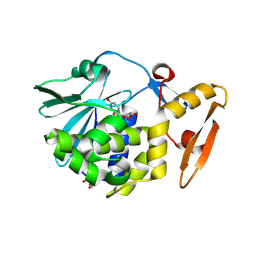 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with Uridine at 1.97 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
5ILX
 
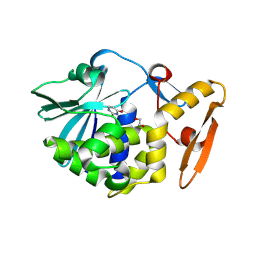 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
8Y9X
 
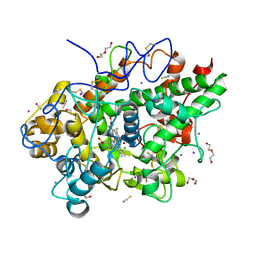 | | Crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Viswanathan, V, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the order of preference of inorganic substrates in mammalian heme peroxidases: crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl
To Be Published
|
|
7XU8
 
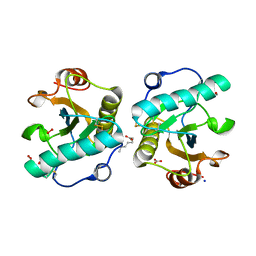 | | Structure of the complex of camel peptidoglycan recognition protein-short (PGRP-S) with heptanoic acid at 2.15 A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Maurya, A, Ahmad, N, Viswanathan, V, Singh, P.K, Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand recognition by peptidoglycan recognition protein-S (PGRP-S): structure of the complex of camel PGRP-S with heptanoic acid at 2.15 angstrom resolution.
Int J Biochem Mol Biol, 13, 2022
|
|
1CL5
 
 | |
6KMK
 
 | | Crystal structure of hydrogen peroxide bound bovine lactoperoxidase at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Singh, P.K, Sirohi, H.V, Bhusan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of hydrogen peroxide bound bovine lactoperoxidase at 2.3 A resolution
To Be Published
|
|
2RD4
 
 | | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 1, Phospholipase A2 isoform 2, ... | | Authors: | Mirza, Z, Kaur, A, Singh, N, Sinha, M, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Design of specific inhibitors of Phospholipase A2: Crystal structure of the complex of phospholipase A2 with pentapeptide Leu-Val-Phe-Phe-Ala at 2.9 A resolution
To be Published
|
|
6M7E
 
 | | Structure of bovine lactoperoxidase with multiple iodide ions in the distaline heme cavity. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maurya, A, Viswanathan, V, Pandey, N, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of bovine lactoperoxidase with multiple iodide ions in the distaline heme cavity.
To Be Published
|
|
8ING
 
 | | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahmad, M.I, Viswanathan, V, Kumar, M, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-03-09 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution
To be published
|
|
1KPM
 
 | | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by Vitamin E and its Implications in Inflammation: Crystal Structure of the Complex Formed between Phospholipase A2 and Vitamin E at 1.8 A Resolution. | | Descriptor: | ACETIC ACID, Phospholipase A2, VITAMIN E | | Authors: | Chandra, V, Jasti, J, Kaur, P, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-01-01 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Evidence of a Specific Inhibition of Phospholipase A2 by alpha-Tocopherol (Vitamin E) and its
Implications in Inflammation: Crystal Structure of the Complex Formed Between Phospholipase A2 and
alpha-Tocopherol at 1.8 A Resolution
J.Mol.Biol., 320, 2002
|
|
1JQ9
 
 | | Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Phe-Leu-Ser-Tyr-Lys at 1.8 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2 | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Complex Formed between a Snake Venom Phospholipase A2 and a Potent Peptide Inhibitor Phe-Leu-Ser-Tyr-Lys at 1.8 A Resolution
J.BIOL.CHEM., 277, 2002
|
|
8HCJ
 
 | | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans | | Descriptor: | CALCIUM ION, Xylan 1,4-beta-xylosidase | | Authors: | Vishwakarma, P, Sachdeva, E, Goyal, A, Ethayathulla, A.S, Das, U, Kaur, P. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans
To Be Published
|
|
1JQ8
 
 | | Design of specific inhibitors of phospholipase A2: Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.0 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2, ... | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of specific peptide inhibitors of phospholipase A2: structure of a complex formed between Russell's viper phospholipase A2 and a designed peptide Leu-Ala-Ile-Tyr-Ser (LAIYS).
ACTA CRYSTALLOGR.,SECT.D, 58, 2002
|
|
6A4Y
 
 | | Crystal structure of bovine lactoperoxidase with partial occupancies of iodide and SCN- ions at the substrate binding site on the distal heme side at 1.92 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of bovine lactoperoxidase with partial occupancies of iodide and SCN- ions at the substrate binding site on the distal heme side at 1.92 A resolution
To Be Published
|
|
3CG9
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-L-rhamnopyranose | | Authors: | Sharma, P, Kaur, A, Singh, N, Sharma, S, Bhushan, A, Pathak, K.M.L, Kaur, P, Singh, T.P. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with methyoxane-2,3,4,5-tetrol at 2.9 A resolution
To be Published
|
|
6A75
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.75 A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-02 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.75 A resolution
To Be Published
|
|
6A7D
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.74 A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-02 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.74 A resolution
To Be Published
|
|
6A6D
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.90A resolution | | Descriptor: | DEPHOSPHO COENZYME A, MAGNESIUM ION, Phosphopantetheine adenylyltransferase, ... | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-27 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.90A resolution
To Be Published
|
|
3CR9
 
 | | Crystal structure of the complex of Lactoferrin with 6-(Hydroxymethyl)oxane-2,3,4,5-tetrol at 3.49 A resolution | | Descriptor: | FE (III) ION, Lactotransferrin, alpha-D-glucopyranose | | Authors: | Mir, R, Kaur, A, Singh, A.K, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structure of the complex of Lactoferrin with 6-(Hydroxymethyl)oxane-2,3,4,5-tetrol at 3.49 A resolution
To be Published
|
|
1MF4
 
 | | Structure-based design of potent and selective inhibitors of phospholipase A2: Crystal structure of the complex formed between phosholipase A2 from Naja Naja sagittifera and a designed peptide inhibitor at 1.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2, VAL-ALA-PHE-ARG-SER | | Authors: | Singh, R.K, Vikram, P, Paramsivam, M, Jabeen, T, Sharma, S, Makker, J, Dey, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-08-09 | | Release date: | 2003-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of specific peptide inhibitors for group I phospholipase A2: structure of a complex formed between phospholipase A2 from Naja naja sagittifera (group I) and a designed peptide inhibitor Val-Ala-Phe-Arg-Ser (VAFRS) at 1.9 A resolution reveals unique features
Biochemistry, 42, 2003
|
|
5ZTJ
 
 | | Crystal Structure of GyraseA C-Terminal Domain from Salmonella typhi at 2.4A Resolution | | Descriptor: | DNA gyrase subunit A | | Authors: | Sachdeva, E, Gupta, D, Tiwari, P, Kaur, G, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The pivot point arginines identified in the beta-pinwheel structure of C-terminal domain from Salmonella Typhi DNA Gyrase A subunit.
Sci Rep, 10, 2020
|
|
5ZXM
 
 | | Crystal Structure of GyraseB N-terminal at 1.93A Resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA gyrase subunit B, ... | | Authors: | Tiwari, P, Gupta, D, Sachdeva, E, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural insights into the transient closed conformation and pH dependent ATPase activity of S.Typhi GyraseB N- terminal domain.
Arch.Biochem.Biophys., 701, 2021
|
|
1OXL
 
 | | INHIBITION OF PHOSPHOLIPASE A2 (PLA2) BY (2-CARBAMOYLMETHYL-5-PROPYL-OCTAHYDRO-INDOL-7-YL)-ACETIC ACID (INDOLE): CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PLA2 FROM RUSSELL'S VIPER AND INDOLE AT 1.8 RESOLUTION | | Descriptor: | (2-CARBAMOYLMETHYL-5-PROPYL-OCTAHYDRO-INDOL-7-YL)ACETIC ACID, CARBONATE ION, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Chandra, V, Balasubramanya, R, Kaur, P, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of the secretory phospholipase A2 from Daboia russelli pulchella with an endogenic indole derivative, 2-carbamoylmethyl-5-propyl-octahydro-indol-7-yl-acetic acid at 1.8 A resolution.
Biochim.Biophys.Acta, 1752, 2005
|
|
4QGD
 
 | | Crystal Structure of the Complex of Phospholipase A2 with Gramine derivative at 1.80 A Resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and binding studies of the complexes of phospholipase A2 with five inhibitors
Biochim.Biophys.Acta, 1854, 2015
|
|
2HD4
 
 | | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution | | Descriptor: | 8-mer Peptide from Lactotransferrin, ACETIC ACID, CALCIUM ION, ... | | Authors: | Prem Kumar, R, Singh, A.K, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-20 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of proteinase K inhibited by a lactoferrin octapeptide Gly-Asp-Glu-Gln-Gly-Glu-Asn-Lys at 2.15 A resolution
To be Published
|
|
