4GP5
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4Q9V
 
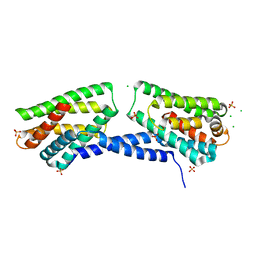 | | Crystal structure of TIPE3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | Authors: | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
4G72
 
 | |
4OBW
 
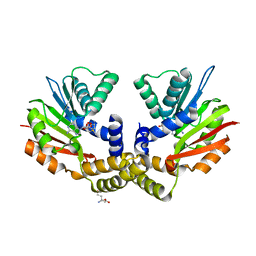 | | crystal structure of yeast Coq5 in the SAM bound form | | Descriptor: | 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial, S-ADENOSYLMETHIONINE, ... | | Authors: | Dai, Y.N, Zhou, K, Cao, D.D, Jiang, Y.L, Meng, F, Chi, C.B, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-01-07 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4I5T
 
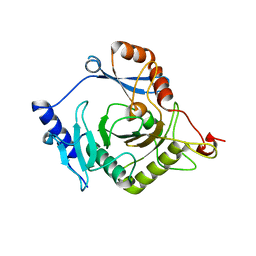 | | Crystal structure of yeast Ap4A phosphorylase Apa2 | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2 | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
3J2G
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2E
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2D
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.700001 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
2LAS
 
 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
3FKB
 
 | | Structure of NDPK H122G and tenofovir-diphosphate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Morera, S, Chen, Y.X. | | Deposit date: | 2008-12-16 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleoside diphosphate kinase and the activation of antiviral phosphonate analogs of nucleotides: binding mode and phosphorylation of tenofovir derivatives
Nucleosides Nucleotides Nucleic Acids, 28, 2009
|
|
3F3R
 
 | | Crystal structure of yeast Thioredoxin1-glutathione mixed disulfide complex | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-1 | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
4HRI
 
 | | Crystal structure of HetR in complex with a 21-bp palindromic DNA at the upstream of the hetP promoter from Anabaena | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Chen, Y, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2012-10-28 | | Release date: | 2013-04-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural and biochemical analyses of Anabaena HetR reveal insights into its binding to DNA targets and the inhibitory hexapeptide ERGSGR
To be Published
|
|
3F3Q
 
 | | Crystal structure of the oxidised form of thioredoxin 1 from saccharomyces cerevisiae | | Descriptor: | Thioredoxin-1, ZINC ION | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
7WF5
 
 | | c-Src in complex with ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Duan, Y, Dai, S, Chen, X, Chen, Y. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural study of ponatinib in inhibiting SRC kinase.
Biochem.Biophys.Res.Commun., 598, 2022
|
|
7WCL
 
 | | Crystal structure of FGFR1 kinase domain with Pemigatinib | | Descriptor: | 11-[2,6-bis(fluoranyl)-3,5-dimethoxy-phenyl]-13-ethyl-4-(morpholin-4-ylmethyl)-5,7,11,13-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),2(6),3,7-tetraen-12-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Lin, Q.M, Jiang, L.Y, Qu, L.Z, Chen, Y.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Characterization of the cholangiocarcinoma drug pemigatinib against FGFR gatekeeper mutants.
Commun Chem, 5, 2022
|
|
7CBY
 
 | | Structure of FOXG1 DNA binding domain bound to DBE2 DNA site | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Dai, S.Y, Li, J, Chen, Y.H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural Basis for DNA Recognition by FOXG1 and the Characterization of Disease-causing FOXG1 Mutations.
J.Mol.Biol., 432, 2020
|
|
6CSV
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 63 kDa,Centrosomal protein of 152 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
3CFU
 
 | | Crystal structure of the yjhA protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR562 | | Descriptor: | Uncharacterized lipoprotein yjhA | | Authors: | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Forouhar, F, Zhao, L, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Swapna, G, Huang, J.Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the yjhA protein from Bacillus subtilis.
To be Published
|
|
6CSU
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 152 kDa, Centrosomal protein of 63 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
6JJ6
 
 | | BRD4 in complex with 500 | | Descriptor: | 1-methyl-6-(3-(4-methylpiperazine-1-carbonyl)benzyl)-1,2a1,5a,6-tetrahydro-2H-pyrido[3',2':6,7]azepino[4,3,2-cd]isoindol-2-one, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | BRD4 in complex with ZZM1
To Be Published
|
|
3QJV
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJR
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
3QJS
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | Descriptor: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | Deposit date: | 2011-01-30 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
6MZ2
 
 | |
8HW4
 
 | | Cryo-EM structure of dehydroepiandrosterone sulfate-bound human ABC transporter ABCC3 in nanodiscs | | Descriptor: | 17-oxoandrost-5-en-3beta-yl hydrogen sulfate, ATP-binding cassette sub-family C member 3 | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
