5U4J
 
 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5UJ3
 
 | | Crystal structure of the KPC-2 beta-lactamase complexed with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2017-01-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis of Substrate Recognition and Product Release by the Klebsiella pneumoniae Carbapenemase (KPC-2).
J. Med. Chem., 60, 2017
|
|
5UJ4
 
 | | Crystal structure of the KPC-2 beta-lactamase complexed with hydrolyzed faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2017-01-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis of Substrate Recognition and Product Release by the Klebsiella pneumoniae Carbapenemase (KPC-2).
J. Med. Chem., 60, 2017
|
|
5UM6
 
 | | Crystal Structure of S. pombe Uba1 in a closed conformation | | Descriptor: | 2-(decylamino)ethane-1-thiol, N-(2-{[(4-chlorophenyl)methyl]disulfanyl}ethyl)decan-1-amine, SULFATE ION, ... | | Authors: | Lv, Z, Yuan, L, Aldana-Masangkay, G, Atkison, J.H, Chen, Y, Olsen, S.K. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Domain alternation and active site remodeling are conserved structural features of ubiquitin E1.
J. Biol. Chem., 292, 2017
|
|
5UJO
 
 | |
5UL8
 
 | |
8JSJ
 
 | |
8JSK
 
 | | Crystal structure of an N-terminal cyclic nucleotide-binding domain of a PycTIR from Pseudovibrio sp. in complex with cUMP | | Descriptor: | PycTIR, Uridine-3',5'-cyclic monophosphate | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-06-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural and functional characterization of cyclic pyrimidine-regulated anti-phage system.
Nat Commun, 15, 2024
|
|
8JSF
 
 | |
8JSZ
 
 | | Crystal structure of a uridylate cyclase from Anabaena sp. | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-06-20 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and functional characterization of cyclic pyrimidine-regulated anti-phage system.
Nat Commun, 15, 2024
|
|
5VLE
 
 | |
5WFI
 
 | |
5TOP
 
 | |
6LR0
 
 | | structure of human bile salt exporter ABCB11 | | Descriptor: | Bile salt export pump | | Authors: | Wang, L, Hou, W.T, Chen, L, Jiang, Y.L, Xu, D, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human bile salts exporter ABCB11.
Cell Res., 30, 2020
|
|
3EH4
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
1SQ4
 
 | | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14 | | Descriptor: | Glyoxylate-induced Protein, THIOCYANATE ION | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14
To be Published
|
|
5WQD
 
 | |
5TOY
 
 | |
3EH3
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
3EH5
 
 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
7LYI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
4R0M
 
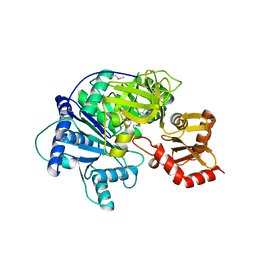 | | Structure of McyG A-PCP complexed with phenylalanyl-adenylate | | Descriptor: | ADENOSINE-5'-[PHENYLALANINYL-PHOSPHATE], McyG protein | | Authors: | Tan, X.F, Dai, Y.N, Zhou, K, Jiang, Y.L, Ren, Y.M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the adenylation-peptidyl carrier protein didomain of the Microcystis aeruginosa microcystin synthetase McyG.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8SPJ
 
 | |
8SXO
 
 | |
