8I7R
 
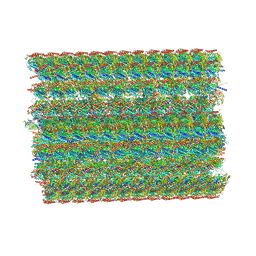 | | In situ structure of axonemal doublet microtubules in mouse sperm with 48-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
8I7O
 
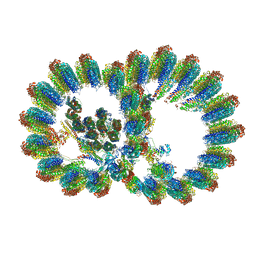 | | In situ structure of axonemal doublet microtubules in mouse sperm with 16-nm repeat | | Descriptor: | Cilia and flagella-associated protein 77, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 276, ... | | Authors: | Zhu, Y, Yin, G.L, Tai, L.H, Sun, F. | | Deposit date: | 2023-02-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In-cell structural insight into the stability of sperm microtubule doublet.
Cell Discov, 9, 2023
|
|
4LWP
 
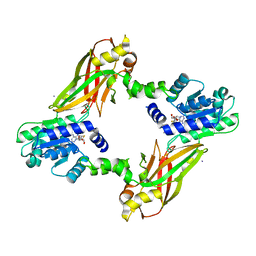 | | Crystal structure of PRMT6-SAH | | Descriptor: | Arginine N-methyltransferase, putative, IODIDE ION, ... | | Authors: | Zhu, Y, Wang, C, Shi, Y, Teng, M. | | Deposit date: | 2013-07-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal Structure of Arginine Methyltransferase 6 from Trypanosoma brucei
Plos One, 9, 2014
|
|
5CMZ
 
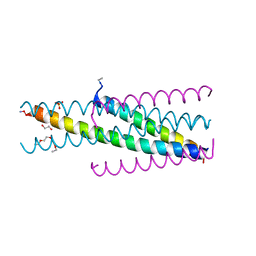 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
4LWO
 
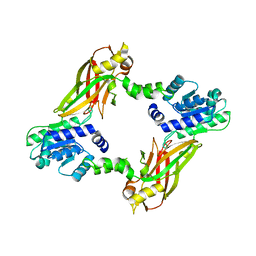 | | Crystal structure of PRMT6 | | Descriptor: | Arginine N-methyltransferase, putative | | Authors: | Zhu, Y, Wang, C, Shi, Y, Teng, M. | | Deposit date: | 2013-07-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Arginine Methyltransferase 6 from Trypanosoma brucei
Plos One, 9, 2014
|
|
5CN0
 
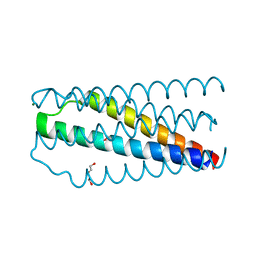 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
5DAY
 
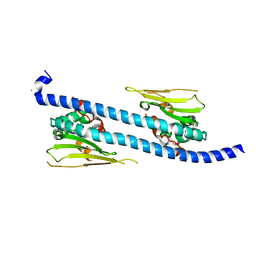 | | The structure of NAP1-Related Protein(NRP1) in Arabidopsis | | Descriptor: | CALCIUM ION, NAP1-related protein 1 | | Authors: | Zhu, Y, Rong, L, Yang, Y, Zhang, C, Feng, H.Y, Zheng, L.N, Shen, W.H, Ma, J.B, Dong, A.W. | | Deposit date: | 2015-08-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | The structure of NAP1-Related Protein(NRP1) in Arabidopsis
To Be Published
|
|
4KDR
 
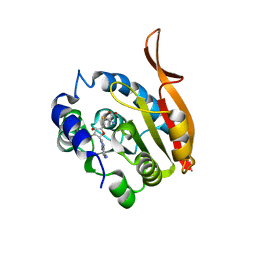 | | Crystal Structure of UBIG/SAH complex | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal Structure of the UBIG/SAH complex
To be Published
|
|
4KDC
 
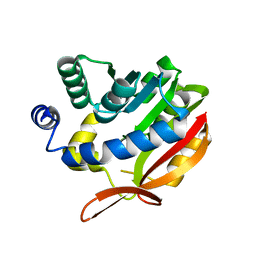 | | Crystal Structure of UBIG | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins.
Biochem. J., 470, 2015
|
|
4KUC
 
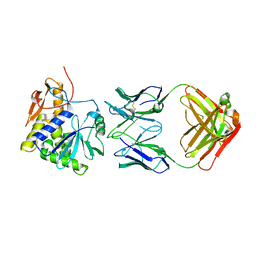 | |
4NJL
 
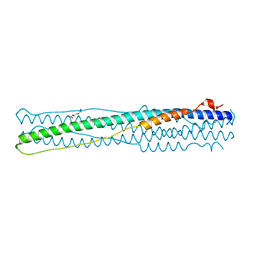 | | Crystal structure of middle east respiratory syndrome coronavirus S2 protein fusion core | | Descriptor: | S protein, TRIETHYLENE GLYCOL | | Authors: | Zhu, Y, Lu, L, Qin, L, Ye, S, Jiang, S, Zhang, R. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor.
Nat Commun, 5, 2014
|
|
5DPM
 
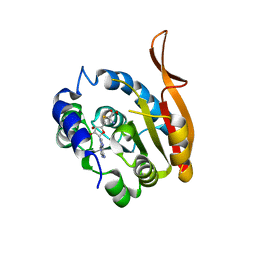 | |
5EQJ
 
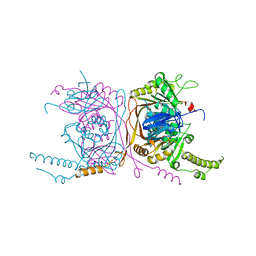 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
5ERG
 
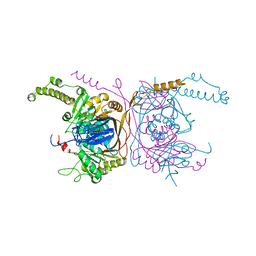 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase TRM6-TRM61 in complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
5H0N
 
 | |
5J8J
 
 | |
5HFM
 
 | | Gp41-targeting HIV-1 fusion inhibitors with hook-like Ile-Asp-Leu tail | | Descriptor: | Envelope glycoprotein gp160,gp41 CHR region, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2016-01-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Rational improvement of gp41-targeting HIV-1 fusion inhibitors: an innovatively designed Ile-Asp-Leu tail with alternative conformations
Sci Rep, 6, 2016
|
|
5HFL
 
 | |
7F01
 
 | |
7FB3
 
 | |
7FB0
 
 | |
7FB1
 
 | |
7FB4
 
 | |
4Q77
 
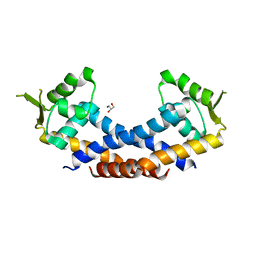 | | Crystal structure of Rot, a global regulator of virulence genes in Staphylococcus aureus | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator rot | | Authors: | Zhu, Y, Fan, X, Li, X, Teng, M. | | Deposit date: | 2014-04-24 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of Rot, a global regulator of virulence genes in Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
