7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | Descriptor: | Reduced folate transporter, cGAMP | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
5WYD
 
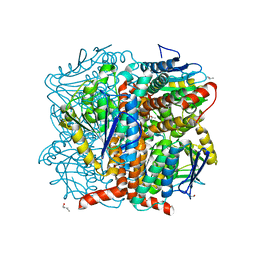 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
8J7S
 
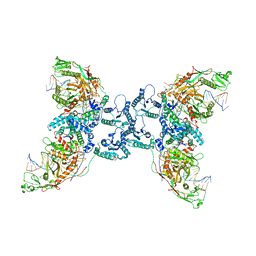 | | Structure of the SPARTA complex | | Descriptor: | DNA (5'-D(P*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*CP*AP*AP*TP*AP*GP*A)-3'), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*A)-3'), ... | | Authors: | Guo, M, Zhu, Y, Lin, Z, Huang, Z. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the ssDNA-activated SPARTA complex.
Cell Res., 33, 2023
|
|
7WSP
 
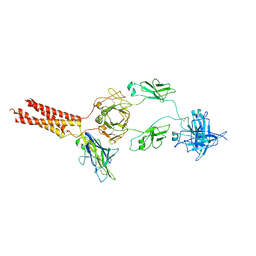 | | Structure of a membrane protein M | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Isoform 2 of Immunoglobulin heavy constant mu | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structure of a membrane protein M
To Be Published
|
|
7XT6
 
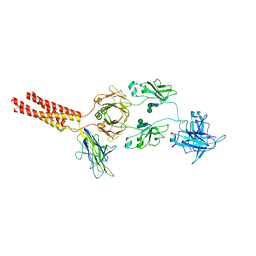 | | Structure of a membrane protein M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, ... | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
8YE6
 
 | |
5WZE
 
 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
8J6R
 
 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6P
 
 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6Q
 
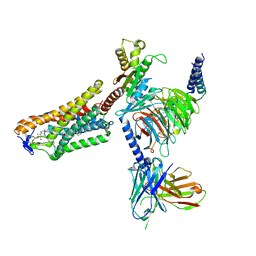 | | Cryo-EM structure of the 3-HB and compound 9n-bound human HCAR2-Gi1 complex | | Descriptor: | (3R)-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8JCN
 
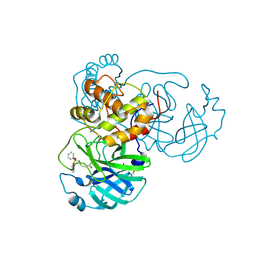 | |
8JCM
 
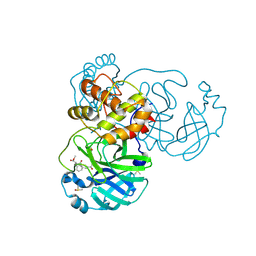 | |
8JCK
 
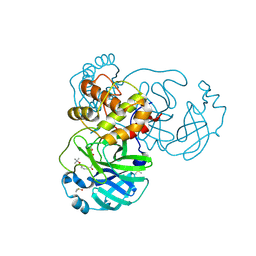 | |
8JCL
 
 | |
8JCO
 
 | |
8JCJ
 
 | |
8IWF
 
 | | Aspergillus niger Rha-2 and pNPR | | Descriptor: | (2S,3R,4R,5R,6S)-2-methyl-6-[4-[oxidanyl(oxidanylidene)-$l^4-azanyl]phenoxy]oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
8IWD
 
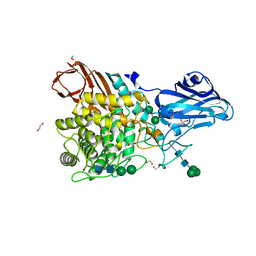 | | Aspergillus niger Rha-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
6VQV
 
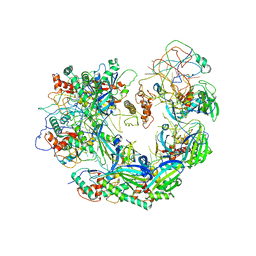 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQW
 
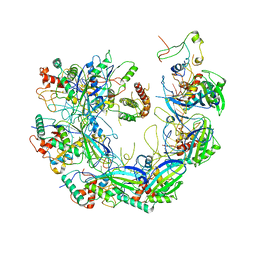 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8WR2
 
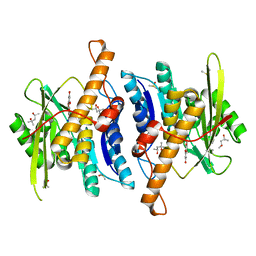 | | Crystal Structure of Human Pyridoxal Kinase with bound Luteolin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Fan, J, Zhu, Y. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and characterization of natural product luteolin as an effective inhibitor of human pyridoxal kinase.
Bioorg.Chem., 143, 2024
|
|
6VQX
 
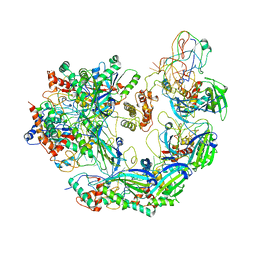 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
