3LZD
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
1NJ9
 
 | | Cocaine hydrolytic antibody 15A10 | | Descriptor: | SODIUM ION, immunoglobulin heavy chain, immunoglobulin variable chain | | Authors: | Larsen, N.A, de Prada, P, Deng, S.X, Zhu, X, Landry, D.W, Wilson, I.A. | | Deposit date: | 2002-12-30 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic and biochemical analysis of cocaine-degrading antibody 15A10.
Biochemistry, 43, 2004
|
|
3LZC
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | Descriptor: | Dph2 | | Authors: | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
5JQG
 
 | | An apo tubulin-RB-TTL complex structure used for side-by-side comparison | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Naismith, J.H, Zhu, X. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule
Nat Commun, 7, 2016
|
|
8SEU
 
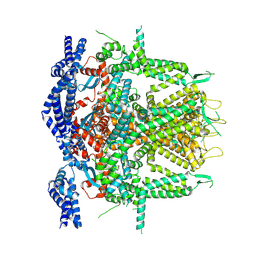 | | Cryo-EM Structure of RyR1 (Local Refinement of TMD) | | Descriptor: | Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SER
 
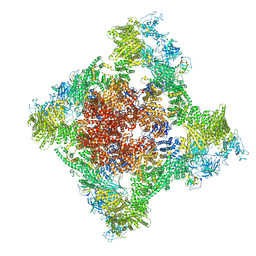 | | Cryo-EM Structure of RyR1 + Adenosine | | Descriptor: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SD4
 
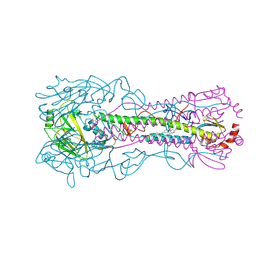 | | Crystal structure of the A/Puerto Rico/8/1934 (H1N1) influenza virus hemagglutinin in complex with small molecule fusion inhibitor compound 7 | | Descriptor: | (S~1~S)-N-{3,5-dichloro-4-[(2S)-2-phenylmorpholine-4-carbonyl]phenyl}-3-[(dimethylamino)methyl]azetidine-1-sulfonimidoyl fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Kadam, R.U, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Ultrapotent influenza hemagglutinin fusion inhibitors developed through SuFEx-enabled high-throughput medicinal chemistry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SEP
 
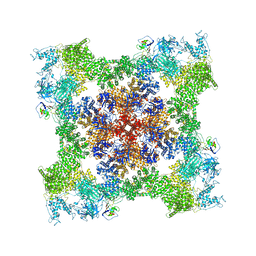 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEY
 
 | | Cryo-EM Structure of RyR1 + Adenosine (Local Refinement of TMD) | | Descriptor: | ADENOSINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SES
 
 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEW
 
 | | Cryo-EM Structure of RyR1 + ADP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SF0
 
 | | Cryo-EM Structure of RyR1 + cAMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SET
 
 | | Cryo-EM Structure of RyR1 + cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEZ
 
 | | Cryo-EM Structure of RyR1 + Adenine (Local Refinement of TMD) | | Descriptor: | ADENINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEN
 
 | | Cryo-EM Structure of RyR1 | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEX
 
 | | Cryo-EM Structure of RyR1 + AMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8TJ4
 
 | | CRYSTAL STRUCTURE OF THE A/Bangkok/1/1979(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ8
 
 | | CRYSTAL STRUCTURE OF THE A/Moscow/10/1999(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ9
 
 | | CRYSTAL STRUCTURE OF THE A/Michigan/15/2014(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ7
 
 | | CRYSTAL STRUCTURE OF THE A/Shandong/9/1993(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJB
 
 | | CRYSTAL STRUCTURE OF THE A/Texas/73/2017(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
8TJ6
 
 | | CRYSTAL STRUCTURE OF THE A/Beijing/353/1989(H3N2) INFLUENZA VIRUS HEMAGGLUTININ WITH HUMAN RECEPTOR ANALOG 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Evolution of human H3N2 influenza virus receptor specificity has substantially expanded the receptor-binding domain site.
Cell Host Microbe, 32, 2024
|
|
