5ZBF
 
 | |
8SUE
 
 | | Human asparagine synthetase (apo-ASNS) | | Descriptor: | Asparagine synthetase [glutamine-hydrolyzing] | | Authors: | Coricello, A, Zhu, W, Lupia, A, Gratteri, C, Vos, M, Chaptal, V, Alcaro, S, Takagi, Y, Richards, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Human asparagine synthetase (apo-ASNS)
To Be Published
|
|
7TFP
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (1S,3S)-3-amino-4-(difluoromethylene)cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-amino-4-(fluoromethyl)cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-06 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
7TEV
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (3S,4R)-3-amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylate | | Descriptor: | (1S,3R,4S)-3-formyl-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
7TED
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (S,E)-3-amino-4-(fluoromethylene)cyclopent-1-ene-1-carboxylate | | Descriptor: | (1S,3R,4S)-3-formyl-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-04 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
4FYG
 
 | | Structural basis for substrate recognition by a novel Legionella phosphoinositide phosphatase | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, SidF, inhibitor of growth family, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FYE
 
 | | Crystal structure of a Legionella phosphoinositide phosphatase, SidF | | Descriptor: | PHOSPHATE ION, SidF, inhibitor of growth family, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FYF
 
 | | Structural basis for substrate recognition by a novel Legionella phosphoinositide phosphatase | | Descriptor: | MERCURY (II) ION, PHOSPHATE ION, SidF, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7LNM
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclopentene-1-carboxylic acid | | Descriptor: | (1~{R},3~{S},4~{R})-3-methyl-4-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Catlin, D, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LON
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (1R,3S,4R)-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-methylcyclohexane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LOM
 
 | | Ornithine Aminotransferase (OAT) soaked with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (3~{S},4~{S})-4-methyl-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, (4~{R})-4-(fluoranylmethyl)-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
3TH8
 
 | | Structure of E. coli undecaprenyl diphosphate synthase complexed with BPH-1063 | | Descriptor: | (2Z)-4-({3-[3-(hexyloxy)phenyl]propyl}amino)-2-hydroxy-4-oxobut-2-enoic acid, Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Zhu, W, Zhang, Y, Oldfield, E. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
6JHB
 
 | |
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
7BDV
 
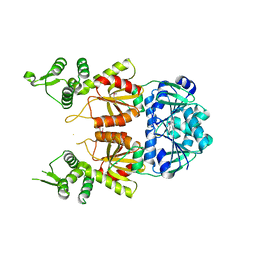 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
8GXR
 
 | |
