2XCF
 
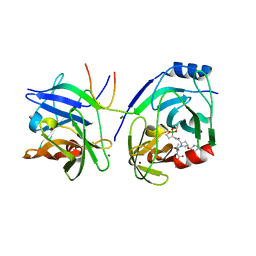 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | CYCLOPENTYL N-[(2S)-1-[(2S,4R)-2-[[(4R)-8-HYDROXY-1,6,10-TRIOXA-5$L^{4}-BORASPIRO[4.5]DECAN-4-YL]CARBAMOYL]-4-ISOQUINOLIN-1-YLOXY-PYRROLIDIN-1-YL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5Y2C
 
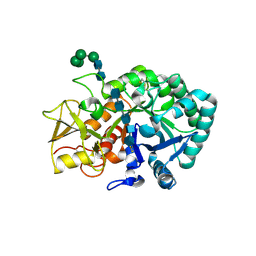 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 2 E2180L mutant in complex with PENTA-N-ACETYLCHITOOCTAOSE (NAG)5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
3AJ2
 
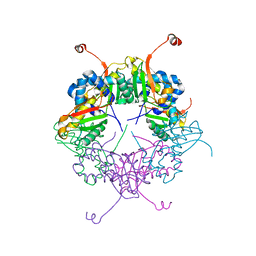 | | The structure of AxCeSD octamer (C-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AJ1
 
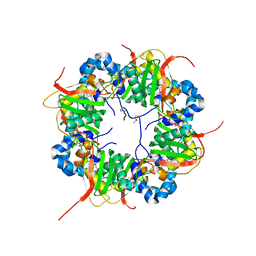 | | The structure of AxCeSD octamer (N-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A8E
 
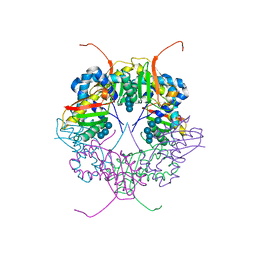 | | The structure of AxCesD octamer complexed with cellopentaose | | Descriptor: | Cellulose synthase operon protein D, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Yao, M, Tanaka, I. | | Deposit date: | 2009-10-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7XAT
 
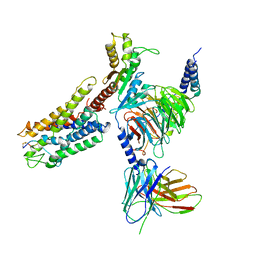 | | Structure of somatostatin receptor 2 bound with SST14. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAU
 
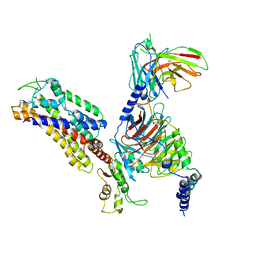 | | Structure of somatostatin receptor 2 bound with octreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7X1U
 
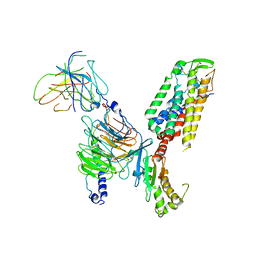 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with an Endogenous Peptide Agonist TRH. | | Descriptor: | Endogenous Peptide Agonist TRH, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
7XAV
 
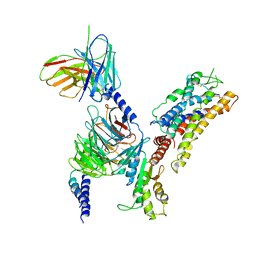 | | Structure of somatostatin receptor 2 bound with lanreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7X1T
 
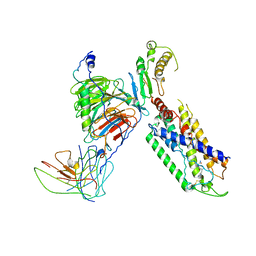 | | Structure of Thyrotropin-Releasing Hormone Receptor bound with Taltirelin. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ScFv16, ... | | Authors: | Yang, F, Zhang, H.H, Meng, X.Y, Li, Y.G, Zhou, Y.X, Ling, S.L, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into thyrotropin-releasing hormone receptor activation by an endogenous peptide agonist or its orally administered analogue.
Cell Res., 32, 2022
|
|
5K6J
 
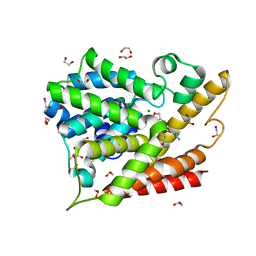 | | Human Phospodiesterase 4B in complex with pyridyloxy-benzoxaborole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-[[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl]oxy]-5-chloranyl-2-(4-oxidanylidenepentoxy)pyridine-3-carbonitrile, MAGNESIUM ION, ... | | Authors: | Rock, F.L, Zhou, Y, Sullivan, D. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | To be published
To Be Published
|
|
8GXN
 
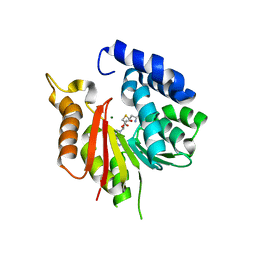 | | The crystal structure of CsFAOMT2 in complex with SAH | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, caffeyl-CoA-O-methyltransferase | | Authors: | Zhang, Z.M, Zhou, Y.E, Huang, H.S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Characterization of two O-methyltransferases involved in the biosynthesis of O-methylated catechins in tea plant.
Nat Commun, 14, 2023
|
|
8GXO
 
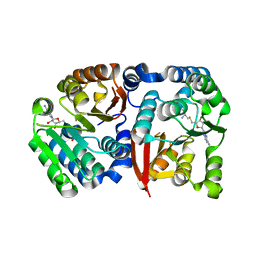 | |
6K2U
 
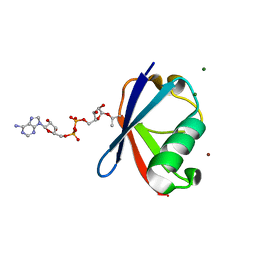 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
7VRG
 
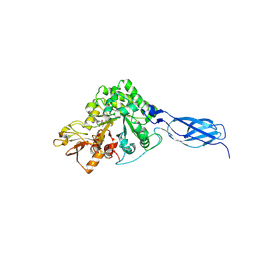 | | Crystal structure of chitinase-h from O. furnacalis in complex with Lynamicin B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Lu, Q, Liu, T, Zhou, Y, Yang, Q. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lynamicin B is a Potential Pesticide by Acting as a Lepidoptera-Exclusive Chitinase Inhibitor.
J.Agric.Food Chem., 69, 2021
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
7VO5
 
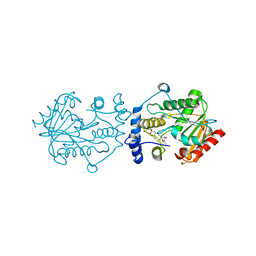 | | Pimaricin type I PKS thioesterase domain (holo Pim TE) | | Descriptor: | (1R,3S,5E,7S,11R,13E,15E,17E,19E,21R,23S,24R,25S)-11,24-dimethyl-1,3,7,21,25-pentakis(oxidanyl)-10,27-dioxabicyclo[21.3.1]heptacosa-5,13,15,17,19-pentaen-9-one, ScnS4 | | Authors: | Bai, L, Zhou, Y. | | Deposit date: | 2021-10-12 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Insights into Chain Release of the Polyene PKS Thioesterase Domain
Acs Catalysis, 12, 2022
|
|
7VO4
 
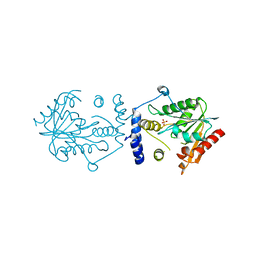 | |
7F5G
 
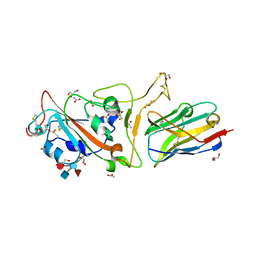 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
5ZNS
 
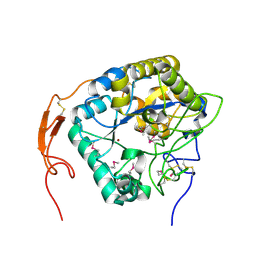 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
5ZNT
 
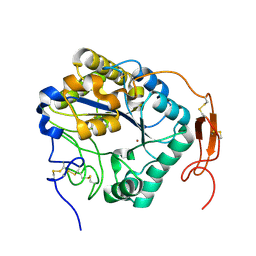 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
6INW
 
 | | A Pericyclic Reaction enzyme | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Feng, Y, Chang, M, Wang, H, Liu, Z, Zhou, Y. | | Deposit date: | 2018-10-28 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6AF0
 
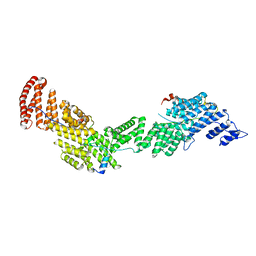 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6L15
 
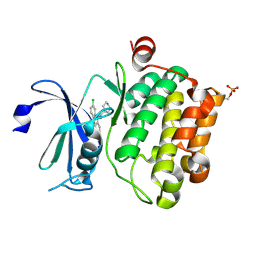 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L12
 
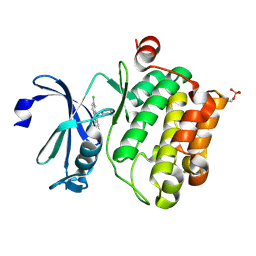 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
