1FWL
 
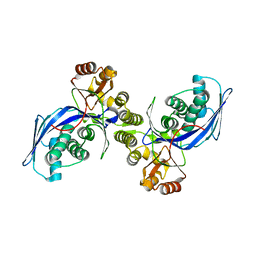 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE | | Descriptor: | HOMOSERINE KINASE | | Authors: | Zhou, T, Daugherty, M, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2000-09-22 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily.
Structure Fold.Des., 8, 2000
|
|
1IHU
 
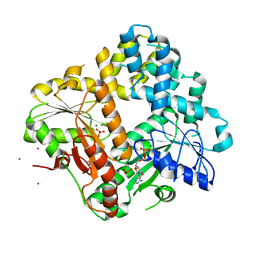 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH MG-ADP-ALF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ARSENICAL PUMP-DRIVING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2001-04-20 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1II9
 
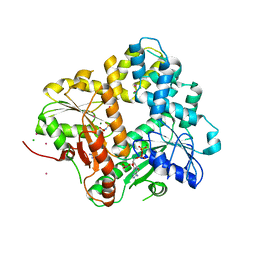 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARSENICAL PUMP-DRIVING ATPASE, CADMIUM ION, ... | | Authors: | Zhou, T, Radaev, S, Gatti, D.L, Rosen, B.P. | | Deposit date: | 2001-04-21 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1II0
 
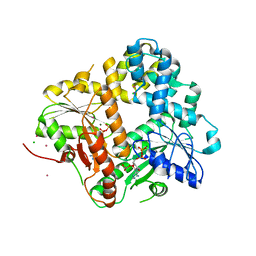 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARSENICAL PUMP-DRIVING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2001-04-20 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
7RI1
 
 | |
7RI2
 
 | |
7R73
 
 | |
7R74
 
 | |
8FBW
 
 | | Crystal structure of SIV-1 V2 antibody NCI05 in complex with a V2 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Kwong, P.D, Van Wazer, D.J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effect of Passive Administration of Monoclonal Antibodies Recognizing Simian Immunodeficiency Virus (SIV) V2 in CH59-Like Coil/Helical or beta-Sheet Conformations on Time of SIV mac251 Acquisition.
J.Virol., 97, 2023
|
|
7MXD
 
 | |
7N28
 
 | |
1KR2
 
 | | CRYSTAL STRUCTURE OF HUMAN NMN/NAMN ADENYLYL TRANSFERASE COMPLEXED WITH TIAZOFURIN ADENINE DINUCLEOTIDE (TAD) | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
1KQN
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
1KQO
 
 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with deamido-NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Hhuman of Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase.
Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2002
|
|
4XMP
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC08 in complex with HIV-1 clade A strain Q842.d12 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-15 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7831 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
4XNZ
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC06B in complex with HIV-1 clade A/E strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.389 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
4XNY
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC08C in complex with HIV-1 clade A strain Q842.d12 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160,Envelope glycoprotein gp160,Envelope glycoprotein gp160, HEAVY CHAIN OF ANTIBODY VRC08C, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
6SZZ
 
 | |
6T00
 
 | |
6URM
 
 | | Crystal structure of vaccine-elicited receptor-binding site targeting antibody LPAF-a.01 in complex with Hemagglutinin H1 A/California/04/2009 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Zhou, T, Cheung, S.F, Kwong, P.D. | | Deposit date: | 2019-10-23 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation.
Cell Rep, 32, 2020
|
|
5JXA
 
 | | Crystal structure of ligand-free VRC03 antigen-binding fragment. | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Zhou, T, Moquin, S, Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Somatic Hypermutation-Induced Changes in the Structure and Dynamics of HIV-1 Broadly Neutralizing Antibodies.
Structure, 24, 2016
|
|
3OY3
 
 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
3OXZ
 
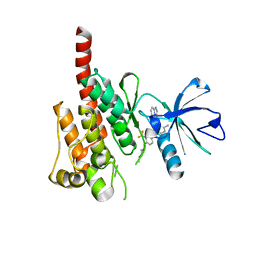 | | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534 | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
4RX4
 
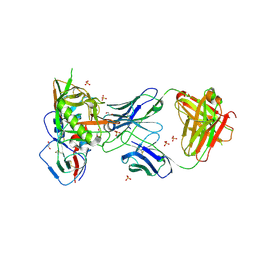 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 8ANC134 in complex with HIV-1 clade A Q842.d12 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC134 Heavy chain, ... | | Authors: | Zhou, T, Acharya, P, Moquin, S, Kwong, P.D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-01 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell(Cambridge,Mass.), 161, 2015
|
|
4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
