2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
4XBX
 
 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBT
 
 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH complexed with (S,S)-cyclohexanediol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, CITRATE ANION, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDV
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclohexanediol | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBY
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDW
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6LCU
 
 | |
8KIF
 
 | | The structure of MmaE with substrate | | Descriptor: | (3R)-3-(2-hydroxy-2-oxoethylamino)decanoic acid, FE (II) ION, Putative dioxygenase | | Authors: | Chen, J, Zhou, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Variation in biosynthesis and metal-binding properties of isonitrile-containing peptides produced by Mycobacteria versus Streptomyces.
Acs Catalysis, 14, 2024
|
|
8KHT
 
 | | The structure of Rv0097 with substrate | | Descriptor: | (3R)-3-(2-hydroxy-2-oxoethylamino)decanoic acid, FE (II) ION, Oxidoreductase | | Authors: | Chen, J, Zhou, J. | | Deposit date: | 2023-08-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Variation in biosynthesis and metal-binding properties of isonitrile-containing peptides produced by Mycobacteria versus Streptomyces.
Acs Catalysis, 14, 2024
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
8JLV
 
 | |
4JFG
 
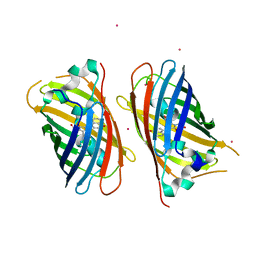 | | Crystal structure of sfGFP-66-HqAla | | Descriptor: | CESIUM ION, Green fluorescent protein, quinolin-8-ol | | Authors: | Wang, J, Liu, X, Li, J, Zhang, W, Hu, M, Zhou, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Significant expansion of the fluorescent protein chromophore through the genetic incorporation of a metal-chelating unnatural amino acid.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
8JN0
 
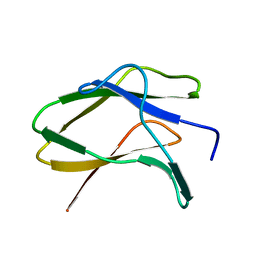 | | N/F domain of alkaline amylase Amy703 | | Descriptor: | Alpha-amylase | | Authors: | Xiang, L, Zhang, G, Zhou, J. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.22826374 Å) | | Cite: | Truncation of N-terminus domain of alkaline a-amylase to form a unique dimer leads to improved activity and stability and decreased calcium ion dependence
To Be Published
|
|
8JNX
 
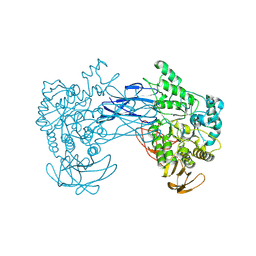 | | alkaline amylase Amy703 with truncated of N-terminus domain | | Descriptor: | Alpha-amylase, CALCIUM ION | | Authors: | Xiang, L, Zhang, G, Zhou, J. | | Deposit date: | 2023-06-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.20279884 Å) | | Cite: | N-terminal domain truncation yielded a unique dimer of polysaccharide hydrolase with enhanced enzymatic activity, stability and calcium ion independence.
Int.J.Biol.Macromol., 266, 2024
|
|
6Z96
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz) soaked with 4-thiouracil (12.65 keV, 7.125 keV (Fe-SAD) and 6.5 keV (S-SAD) data) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, HYDROSULFURIC ACID, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.327 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z92
 
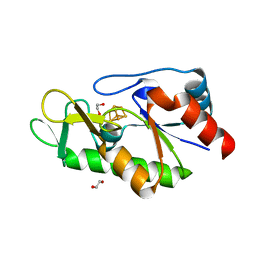 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz) solved by Fe-SAD phasing | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DUF523 domain-containing protein, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z93
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z94
 
 | | [4Fe-4S]-dependent thiouracil desulfidase TudS (DUF523Vcz)(S-SAD data) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DUF523 domain-containing protein, ... | | Authors: | Pecqueur, L, Zhou, J, Fontecave, M, Golinelli-Pimpaneau, B. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Structural Evidence for a [4Fe-5S] Intermediate in the Non-Redox Desulfuration of Thiouracil.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
1WNO
 
 | | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Hu, H, Wang, G, Yang, H, Zhou, J, Mo, L, Yang, K, Jin, C, Jin, C, Rao, Z. | | Deposit date: | 2004-08-07 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407
To be Published
|
|
2RKK
 
 | |
2RKL
 
 | | Crystal Structure of S.cerevisiae Vta1 C-terminal domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Xiao, J, Xia, H, Zhou, J, Xu, Z. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of vta1 function in the multivesicular body sorting pathway.
Dev.Cell, 14, 2008
|
|
4RNC
 
 | | Crystal structure of an esterase RhEst1 from Rhodococcus sp. ECU1013 | | Descriptor: | Esterase, PHOSPHATE ION | | Authors: | Dou, S, Kong, X.D, Xu, J.H, Zhou, J. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate channel evolution of an esterase for the synthesis of Cilastatin
CATALYSIS SCIENCE AND TECHNOLOGY, 5, 2015
|
|
1XHL
 
 | | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase family member (5L265), ... | | Authors: | Schormann, N, Karpova, E, Zhou, J, Zhang, Y, Symersky, J, Bunzel, R, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKInstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate
To be Published
|
|
1XKQ
 
 | | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain reductase family member (5D234) | | Authors: | Schormann, N, Zhou, J, Karpova, E, Zhang, Y, Symersky, J, Bunzel, B, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKinstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor
To be Published
|
|
