4OU5
 
 | | Crystal structure of esterase rPPE mutant S159A/W187H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | Authors: | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | Deposit date: | 2014-02-15 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | Descriptor: | B-agarase | | Authors: | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
3HJW
 
 | | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(FHU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, POTASSIUM ION, ... | | Authors: | Liang, B, Zhou, J, Kahen, E, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Nat.Struct.Mol.Biol., 16, 2009
|
|
7VOC
 
 | | The crystal structure of a Radical SAM Enzyme BlsE involved in the Biosynthesis of Blasticidin S | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{R})-6-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Cytosylglucuronate decarboxylase, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.62005424 Å) | | Cite: | Radical S -Adenosyl Methionine Enzyme BlsE Catalyzes a Radical-Mediated 1,2-Diol Dehydration during the Biosynthesis of Blasticidin S.
J.Am.Chem.Soc., 144, 2022
|
|
7W5K
 
 | | The C296A mutant of L-sorbosone dehydrogenase (SNDH) from Gluconobacter Oxydans WSH-004 | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
4P5A
 
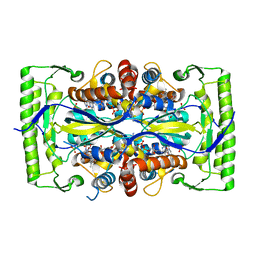 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | Descriptor: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
4P5B
 
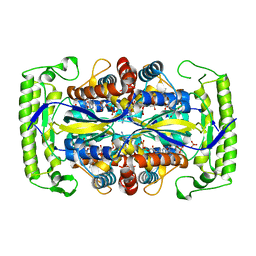 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br dUMP | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Crystal structure of a UMP/dUMP methylase PolB form Streptomyces cacaoi bound with 5-Br dUMP
To Be Published
|
|
7X0B
 
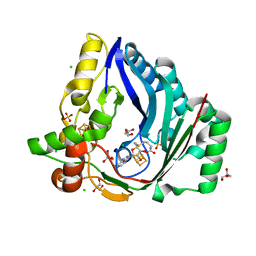 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZX
 
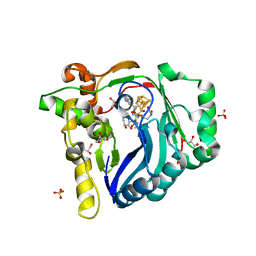 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
5XM6
 
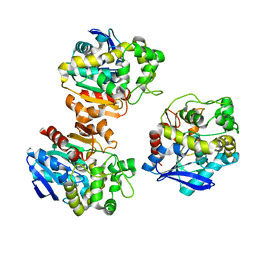 | | the overall structure of VrEH2 | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Kong, X.D, Yu, H.L, Shang, Y.P, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
5X5R
 
 | |
5XMD
 
 | |
5Y5D
 
 | |
7VYO
 
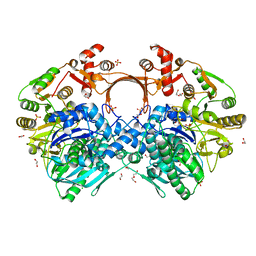 | | The structure of GdmN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
5Y6Y
 
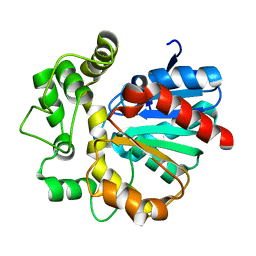 | | The crystal structure of VrEH2 mutant M263N | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Yu, H.L, Chen, Q, Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-08-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
5ZOA
 
 | |
6J1Q
 
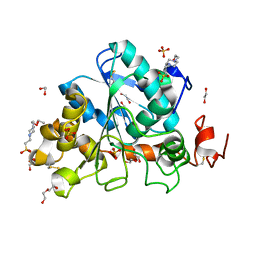 | | Crystal structure of Candida Antarctica Lipase B mutant - RS | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1S
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SS | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lipase B, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1T
 
 | | Crystal structure of Candida Antarctica Lipase B mutant SR with product analogue | | Descriptor: | (2S)-2-phenyl-N-[(1R)-1-phenylethyl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1P
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
6J1R
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - RR | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lipase B, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
8HKS
 
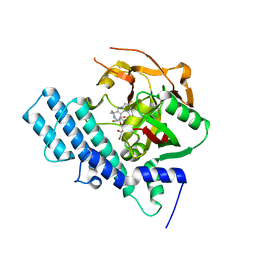 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib(BGB-290) | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib
To Be Published
|
|
8HKO
 
 | |
8HLR
 
 | |
7W5N
 
 | | The crystal structure of the reduced form of Gluconobacter oxydans WSH-004 SNDH | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Yin, D.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
