8DK2
 
 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetABC in an unclamped state trapped in ATP dependent dimeric form | | Descriptor: | DNA (26-MER), JetA, JetB, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
8DK3
 
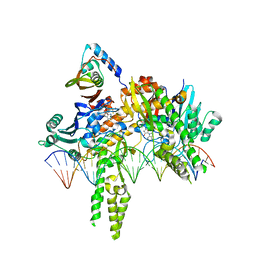 | | CryoEM structure of Pseudomonas aeruginosa PA14 JetC ATPase domain bound to DNA and cWHD domain of JetA | | Descriptor: | DNA (26-MER), JetA, JetC, ... | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
8DK1
 
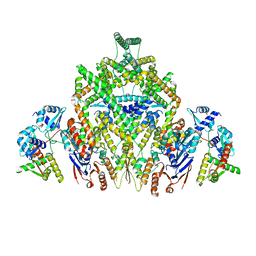 | | CryoEM structure of JetABC (head construct) from Pseudomonas aeruginosa PA14 | | Descriptor: | JetA, JetB, JetC | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-07-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
4Y25
 
 | | Bacterial polysaccharide outer membrane secretin | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine export protein | | Authors: | Wang, Y, AndolePannuri, A, Ni, D, Zhou, H, Cao, X, Lu, X, Romeo, T, Huang, Y. | | Deposit date: | 2015-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structural Basis for Translocation of a Biofilm-supporting Exopolysaccharide across the Bacterial Outer Membrane
J.Biol.Chem., 291, 2016
|
|
4ZRI
 
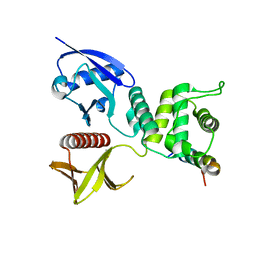 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
8DQL
 
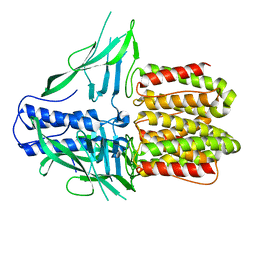 | | CryoEM structure of IglD | | Descriptor: | Secretion system protein | | Authors: | Liu, X, Clemens, D, Lee, B, Yang, X, Zhou, H, Horwitz, M. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic Structure of IglD Demonstrates Its Role as a Component of the Baseplate Complex of the Francisella Type VI Secretion System.
Mbio, 13, 2022
|
|
2L4A
 
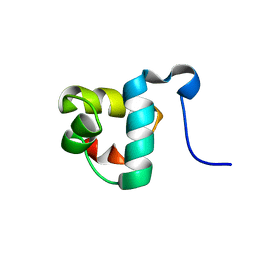 | |
2LD6
 
 | |
4L36
 
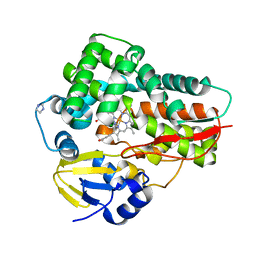 | | Crystal structure of the cytochrome P450 enzyme TxtE | | Descriptor: | HEME B/C, IMIDAZOLE, Putative P450-like protein | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Wang, Z.J, Zhou, H, Yang, M, Chen, Y.X, Tang, L, He, J.H. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Insights into the Mechanism for Recognizing Substrate of the Cytochrome P450 Enzyme TxtE.
Plos One, 8, 2013
|
|
2LP4
 
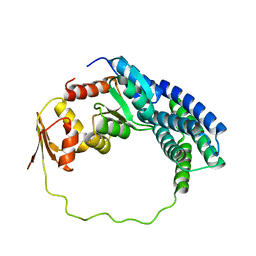 | | Solution structure of P1-CheY/P2 complex in bacterial chemotaxis | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheY | | Authors: | Dahlquist, F, Mo, G, Zhou, H, Kamamura, T. | | Deposit date: | 2012-01-31 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a complex of the histidine autokinase CheA with its substrate CheY.
Biochemistry, 51, 2012
|
|
8YA5
 
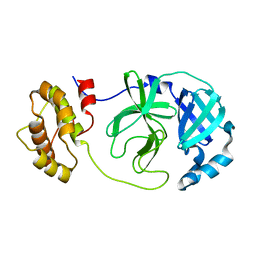 | |
8YA1
 
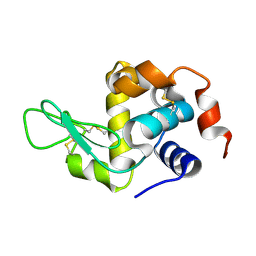 | | HEN EGG WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
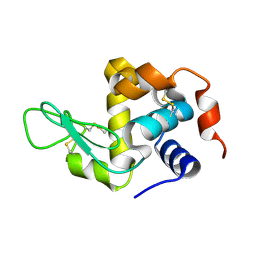
8YA4
 
 | |
8Y9W
 
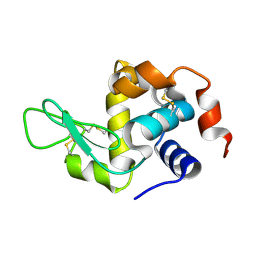 | |
8Y9V
 
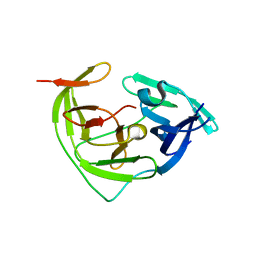 | | ZIKV NS2B/NS3 protease | | Descriptor: | DAR-LYS-ORN-ARG, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H, Wang, Q.S. | | Deposit date: | 2024-02-07 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
2MMU
 
 | | Structure of CrgA, a Cell Division Structural and Regulatory Protein from Mycobacterium tuberculosis, in Lipid Bilayers | | Descriptor: | Cell division protein CrgA | | Authors: | Das, N, Dai, J, Hung, I, Rajagopalan, M, Zhou, H, Cross, T.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MC7
 
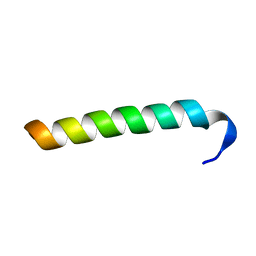 | | Structure of Salmonella MgtR | | Descriptor: | Regulatory peptide | | Authors: | Jean-Francois, F, Dai, J, Yu, L, Myrick, A, Rubin, E, Fajer, P, Song, L, Zhou, H, Cross, T. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Binding of MgtR, a Salmonella Transmembrane Regulatory Peptide, to MgtC, a Mycobacterium tuberculosis Virulence Factor: A Structural Study.
J.Mol.Biol., 426, 2014
|
|
5KWQ
 
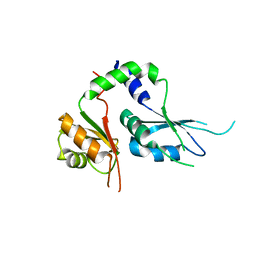 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
1GMY
 
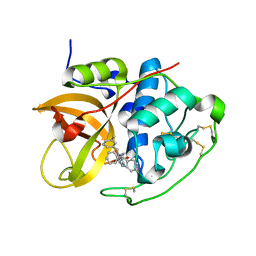 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
1NK1
 
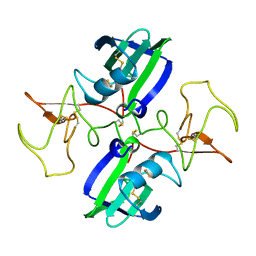 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1K0S
 
 | | Solution structure of the chemotaxis protein CheW from the thermophilic organism Thermotoga maritima | | Descriptor: | CHEMOTAXIS PROTEIN CHEW | | Authors: | Griswold, I.J, Zhou, H, Swanson, R.V, Simon, M.I, Dahlquist, F.W. | | Deposit date: | 2001-09-20 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and interactions of CheW from Thermotoga maritima.
Nat.Struct.Biol., 9, 2002
|
|
7MD5
 
 | | Insulin receptor ectodomain dimer complexed with two IRPA-9 partial agonists | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
7MD4
 
 | | Insulin receptor ectodomain dimer complexed with two IRPA-3 partial agonists | | Descriptor: | Insulin B chain, Insulin chain A, Isoform Short of Insulin receptor, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
8WWQ
 
 | | geniposidic acid O-methyltransferase complexed with SAH and geniposidic acid | | Descriptor: | 1,2-ETHANEDIOL, Geniposidic acid, MAGNESIUM ION, ... | | Authors: | Luo, Z, Li, L, Ma, D, Zhou, H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the substrate specificity of GAMT and divergence of seco-iridoid and closed-ring iridoid
To Be Published
|
|
7X25
 
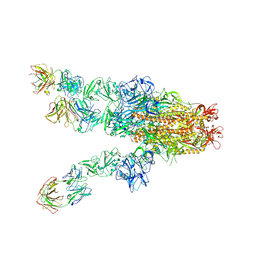 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class4 (2u1d RBD with 3Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J, Zhang, S, Zhou, H, Wang, X. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
