7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7ZJV
 
 | |
8WKU
 
 | | Complex structure of MjHKU4r-CoV-1 spike RBD bound to human CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form, MjHKU4r-CoV-1 spike RBD, ... | | Authors: | Zhao, Z.N, Chai, Y, Gao, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for receptor recognition and broad host tropism for merbecovirus MjHKU4r-CoV-1.
Embo Rep., 25, 2024
|
|
8HIF
 
 | | One asymmetric unit of Singapore grouper iridovirus capsid | | Descriptor: | Major capsid protein, Penton protein (VP14), VP137, ... | | Authors: | Zhao, Z.N, Liu, C.C, Zhu, D.J, Qi, J.X, Zhang, X.Z, Gao, G.F. | | Deposit date: | 2022-11-20 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic architecture of Singapore grouper iridovirus and implications for giant virus assembly.
Nat Commun, 14, 2023
|
|
2Q8O
 
 | |
4A82
 
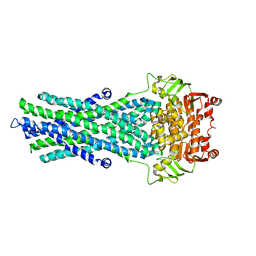 | | Fitted model of staphylococcus aureus sav1866 model ABC transporter in the human cystic fibrosis transmembrane conductance regulator volume map EMD-1966. | | Descriptor: | CYSTIC FIBROSIS TRANSMEMBRANE CONDUCTANCE REGULATOR | | Authors: | Rosenberg, M.F, ORyan, L.P, Hughes, G, Zhao, Z, Aleksandrov, L.A, Riordan, J.R, Ford, R.C. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | The Cystic Fibrosis Transmembrane Conductance Regulator (Cftr):3D Structure and Localisation of a Channel Gate.
J.Biol.Chem., 286, 2011
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
8KA8
 
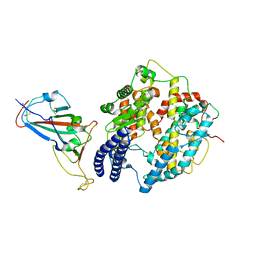 | | Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8KC2
 
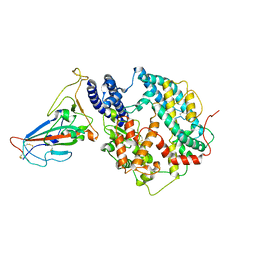 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8K1I
 
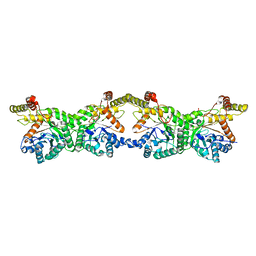 | |
3PS5
 
 | | Crystal structure of the full-length Human Protein Tyrosine Phosphatase SHP-1 | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, W, Liu, L, Song, X, Mo, Y, Komma, C, Bellamy, H.D, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation.
J.Cell.Biochem., 112, 2011
|
|
8K9H
 
 | |
6IQD
 
 | | Crystal structure of Alcohol dehydrogenase from Geobacillus stearothermophilus | | Descriptor: | Alcohol dehydrogenase, ZINC ION | | Authors: | Xue, S, Feng, Y, Guo, X, Zhao, Z. | | Deposit date: | 2018-11-07 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Characterization of the substrate scope of an alcohol dehydrogenase commonly used as methanol dehydrogenase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6JX1
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | Descriptor: | Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUK
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JWG
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
1GWZ
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
8YE3
 
 | | Cryo-EM structure of human respiratory syncytial virus fusion protein variant | | Descriptor: | Fusion glycoprotein F0 | | Authors: | Li, Q.M, Su, J.G, Zhang, J, Liang, Y, Shao, S, Li, X.Y, Zhao, Z.X. | | Deposit date: | 2024-02-21 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mutating a flexible region of the RSV F protein can stabilize the prefusion conformation.
Science, 385, 2024
|
|
5J3J
 
 | | Crystal Structure of human DPP-IV in complex with HL1 | | Descriptor: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
4C7A
 
 | | Crystal structure of the Smoothened CRD, selenomethionine-labeled | | Descriptor: | SMOOTHENED, SODIUM ION, ZINC ION | | Authors: | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
4C79
 
 | | Crystal structure of the Smoothened CRD, native | | Descriptor: | SMOOTHENED, SODIUM ION, ZINC ION | | Authors: | Nachtergaele, S, Whalen, D.M, Mydock, L.K, Zhao, Z, Malinauskas, T, Krishnan, K, Ingham, P.W, Covey, D.F, Rohatgi, R, Siebold, C. | | Deposit date: | 2013-09-20 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structure and Function of the Smoothened Extracellular Domain in Vertebrate Hedgehog Signaling
Elife, 2, 2013
|
|
6IH3
 
 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH5
 
 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
 | |
