6ILM
 
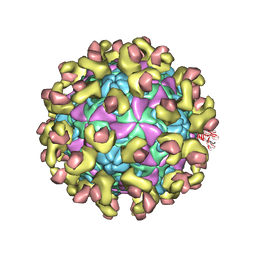 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 7.4 | | Descriptor: | Beta-2-microglobulin, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILN
 
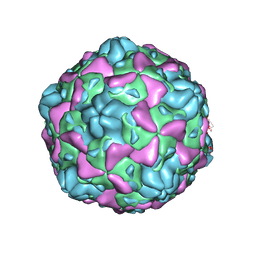 | | Cryo-EM structure of full Echovirus 6 particle at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
5XZ3
 
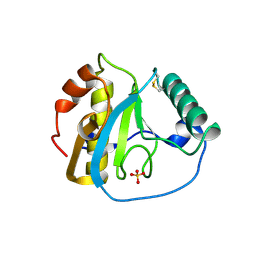 | | The X-ray structure of Apis mellifera PGRP-SA | | Descriptor: | Peptidoglycan-recognition protein, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae).
Protein Sci., 27, 2018
|
|
6ILK
 
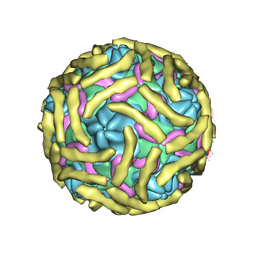 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILP
 
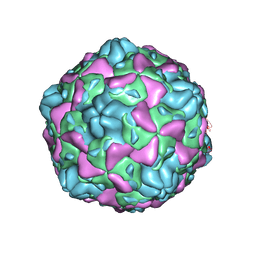 | | Cryo-EM structure of full Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILL
 
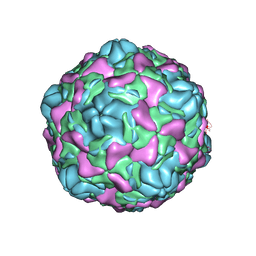 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
2BBO
 
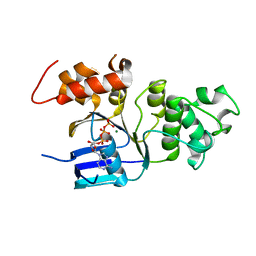 | | Human NBD1 with Phe508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
6ILO
 
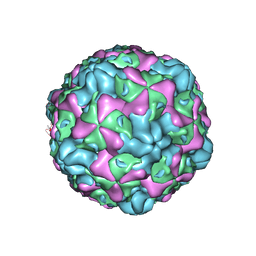 | | Cryo-EM structure of empty Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
2BBS
 
 | | Human deltaF508 NBD1 with three solubilizing mutations | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
2BBT
 
 | | Human deltaF508 NBD1 with two solublizing mutations. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
1VE7
 
 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 in complex with p-nitrophenyl phosphate | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
8V0C
 
 | | Structure of TDP1 catalytic domain complexed with compound IB06 | | Descriptor: | (8M)-8-(2-{[2-(fluorosulfonyl)ethyl]amino}phenyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of TDP1 complexed with inhibitors
To Be Published
|
|
8UZZ
 
 | | Structure of TDP1 catalytic domain complexed with compound IB03 | | Descriptor: | (8M)-8-{2-[(fluorosulfonyl)oxy]phenyl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structureal analysis of TDP1 in complex with inhbitors
To Be Published
|
|
8V0B
 
 | | Structure of TDP1 catalytic domain complexed with compound IB05 | | Descriptor: | 1,2-ETHANEDIOL, 8-{4-[(fluorosulfonyl)oxy]phenyl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of TDP1 complexed with inhibitors
To Be Published
|
|
8UV1
 
 | | Structure of TDP1 complexed with compound IB01 | | Descriptor: | 1,2-ETHANEDIOL, 8-(fluorosulfonyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke, T.R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of TDP1 complexed with compound IB01
To Be Published
|
|
8UZV
 
 | | Structure of TDP1 catalytic domain complexed with compound IB02 | | Descriptor: | 1,2-ETHANEDIOL, 8-{[2-(fluorosulfonyl)ethyl]amino}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Structure of TDP1 catalytic domain complexed with compound IB02
To Be Published
|
|
8WTI
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
1DKX
 
 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 1 SELENOMETHIONYL CRYSTALS | | Descriptor: | SUBSTRATE BINDING DOMAIN OF DNAK, SUBSTRATE PEPTIDE (7 RESIDUES) | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
1DKY
 
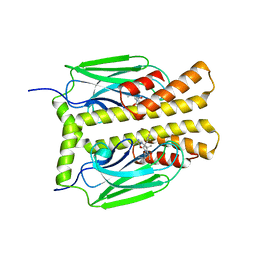 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 2 NATIVE CRYSTALS | | Descriptor: | DNAK, PEPTIDE SUBSTRATE | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
1DKZ
 
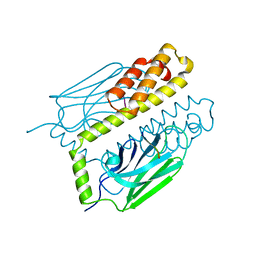 | | THE SUBSTRATE BINDING DOMAIN OF DNAK IN COMPLEX WITH A SUBSTRATE PEPTIDE, DETERMINED FROM TYPE 1 NATIVE CRYSTALS | | Descriptor: | SUBSTRATE BINDING DOMAIN OF DNAK, SUBSTRATE PEPTIDE (7 RESIDUES) | | Authors: | Zhu, X, Zhao, X, Burkholder, W.F, Gragerov, A, Ogata, C.M, Gottesman, M.E, Hendrickson, W.A. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of substrate binding by the molecular chaperone DnaK.
Science, 272, 1996
|
|
2D3D
 
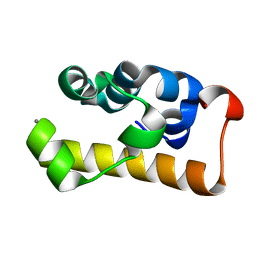 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
1OWH
 
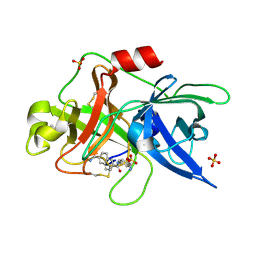 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
8WYG
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 2 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
1OWE
 
 | | Substituted 2-Naphthamidine inhibitors of urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-PHENYL-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Rockway, T.W, Geyer, A, McClellan, W, Weitzberg, M, Zhao, X, Mantei, R, Nienaber, V.L, Stewart, K, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2003-03-28 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of Novel Binding Interactions in the Development of Potent, Selective 2-Naphthamidine Inhibitors of Urokinase. Synthesis, Structural Analysis, and SAR of N-Phenyl Amide 6-Substitution.
J.Med.Chem., 47, 2004
|
|
8WY7
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
