1RCS
 
 | | NMR STUDY OF TRP REPRESSOR-OPERATOR DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*AP*CP*G)-3'), TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the trp repressor-operator DNA complex.
J.Mol.Biol., 238, 1994
|
|
4W5A
 
 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1WRT
 
 | | NMR STUDY OF APO TRP REPRESSOR | | Descriptor: | APO TRP REPRESSOR | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
1WRS
 
 | | NMR STUDY OF HOLO TRP REPRESSOR | | Descriptor: | HOLO TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
1A3Z
 
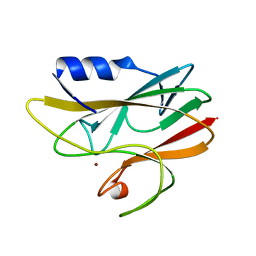 | | REDUCED RUSTICYANIN AT 1.9 ANGSTROMS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Zhao, D, Shoham, M. | | Deposit date: | 1998-01-27 | | Release date: | 1998-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rusticyanin: Extremes in acid stability and redox potential explained by the crystal structure.
Biophys.J., 74, 1998
|
|
2MOX
 
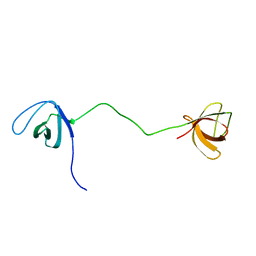 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
4LNP
 
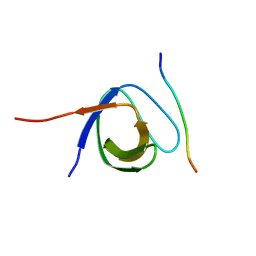 | | The first SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, Vinculin | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
4LN2
 
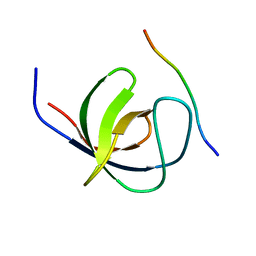 | | The second SH3 domain from CAP/Ponsin in complex with proline rich peptide from Vinculin | | Descriptor: | Sorbin and SH3 domain-containing protein 1, proline rich peptide | | Authors: | Zhao, D, Li, F, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2013-07-11 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
7VBA
 
 | | Structure of the pre state human RNA Polymerase I Elongation Complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(P*A*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*AP*GP*AP*GP*AP*CP*AP*GP*CP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBC
 
 | | Back track state of human RNA Polymerase I Elongation Complex | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*AP*GP*CP*GP*TP*GP*TP*CP*AP*GP*CP*AP*AP*TP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBB
 
 | | Structure of the post state human RNA Polymerase I Elongation Complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*CP*GP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
5IQL
 
 | |
5XTZ
 
 | | Crystal structure of GAS41 YEATS bound to H3K27ac peptide | | Descriptor: | ACETATE ION, THR-LYS-ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, YEATS domain-containing protein 4 | | Authors: | Li, H.T, Zhao, D. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity.
Cell Discov, 4, 2018
|
|
5HJB
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K9 | | Descriptor: | Protein AF-9, peptide of Histone H3.1 | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-12 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
5HJD
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K18 | | Descriptor: | COPPER (II) ION, Protein AF-9, SULFATE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain.
Mol.Cell, 62, 2016
|
|
5HJC
 
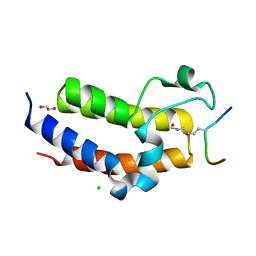 | | BRD3 second bromodomain in complex with histone H3 acetylation at K18 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, CHLORIDE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
5XNV
 
 | | Crystal structure of YEATS2 YEATS bound to H3K27ac peptide | | Descriptor: | ALA-ALA-ARG-ALY-SER-ALA-PRO-ALA, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Li, H.T, Guan, H.P, Zhao, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer.
Nat Commun, 8, 2017
|
|
8SMX
 
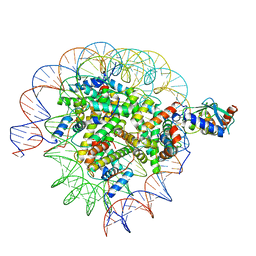 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SMW
 
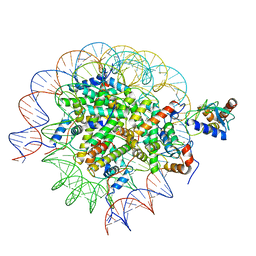 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN6
 
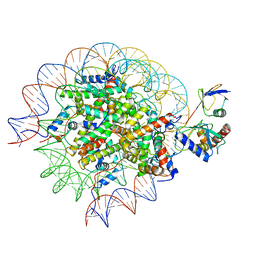 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 4) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN7
 
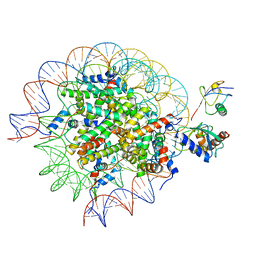 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 5) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN4
 
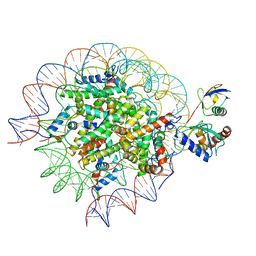 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SMY
 
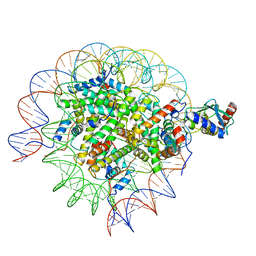 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN2
 
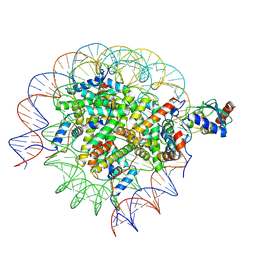 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c (UbcH5c chemically conjugated to histone H2A) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN5
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
