7E7L
 
 | | The crystal structure of arylacetate decarboxylase from Olsenella scatoligenes. | | Descriptor: | 4-HYDROXYPHENYLACETATE, Hydroxyphenylacetic acid decarboxylase | | Authors: | Lu, Q, Duan, Y, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The Glycyl Radical Enzyme Arylacetate Decarboxylase from Olsenella scatoligenes
Acs Catalysis, 11, 2021
|
|
7CQ7
 
 | | Structure of the human CLCN7-OSTM1 complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ADP
To Be Published
|
|
7CQ5
 
 | | Structure of the human CLCN7-OSTM1 complex with ATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ATP
To Be Published
|
|
7CQ6
 
 | | Structure of the human CLCN7-OSTM1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, H(+)/Cl(-) exchange transporter 7, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex
To Be Published
|
|
8H8F
 
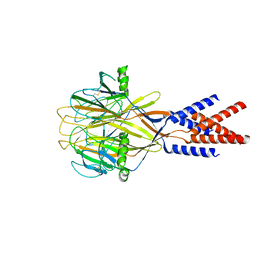 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR (resting state)
To Be Published
|
|
8H8E
 
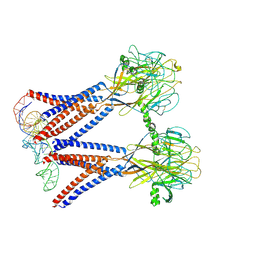 | | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state) | | Descriptor: | Proton-activated chloride channel, tRNA (75-MER)of Spodoptera frugiperda | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the dimeric Xenopus tropical acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (closed state)
To Be Published
|
|
8H8D
 
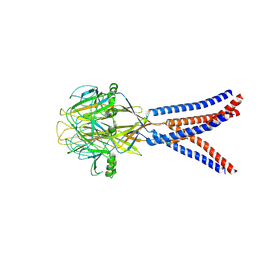 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state)
To Be Published
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
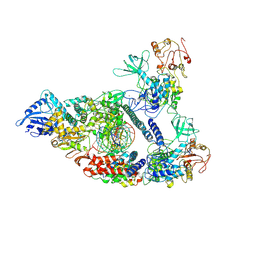 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7C5Z
 
 | |
7DIE
 
 | | Crystal structure of M. penetrans Ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, w.m, Zhang, y, Wang, h.f. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ferritin with Atypical Ferroxidase Centers Takes B-Channels as the Pathway for Fe 2+ Uptake from Mycoplasma .
Inorg.Chem., 60, 2021
|
|
7EGS
 
 | | The crystal structure of lobe domain of E. coli RNA polymerase complexed with the C-terminal domain of UvrD | | Descriptor: | DNA helicase II, DNA-directed RNA polymerase subunit beta, GLYCEROL | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
7FEC
 
 | |
7FED
 
 | |
7FEB
 
 | |
7CKQ
 
 | |
2CEU
 
 | | Despentapeptide insulin in acetic acid (pH 2) | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Zhang, Y, Zakova, L, Dodson, E.J, Turkenburg, J.P, Brange, J, Dodson, G.G. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | I222 Crystal Form of Despentapeptide (B26-B30) Insulin Provides New Insights Into the Properties of Monomeric Insulin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7F65
 
 | | Bacetrial Cocaine Esterase with mutations T172R/G173Q/V116K/S117A/A51L, bound to benzoic acid | | Descriptor: | BENZOIC ACID, Cocaine esterase, SULFATE ION | | Authors: | Ouyang, P.F, Zhang, Y, Tong, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Computational Design and Crystal Structure of a Highly Efficient Benzoylecgonine Hydrolase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7VLZ
 
 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 | | Descriptor: | CALCIUM ION, Peptide P1, Peptide P2, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7
To Be Published
|
|
7F0R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
4B9D
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
8QRH
 
 | | Inactivated tick-borne encephalitis virus (TBEV) vaccine strain Sofjin-Chumakov | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, Small envelope protein M | | Authors: | Moiseenko, A.V, Zhang, Y, Vorovitch, M, Ivanova, A, Liu, Z, Osolodkin, D.I, Egorov, A, Ishmukhametov, A, Sokolova, O.S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The structure of inactivated mature tick-borne encephalitis virus at 3.0 angstrom resolution.
Emerg Microbes Infect, 13, 2024
|
|
3PZ8
 
 | |
7EGT
 
 | | The crystal structure of the C-terminal domain of T. thermophilus UvrD complexed with the N-terminal domain of UvrB | | Descriptor: | DNA helicase UvrD, UvrABC system protein B | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
5TUR
 
 | | Pim-1 kinase in complex with a 7-azaindole | | Descriptor: | 1-methyl-2-[4-(piperazin-1-yl)phenyl]-1H-pyrrolo[2,3-b]pyridine-4-carbonitrile, Serine/threonine-protein kinase pim-1 | | Authors: | Mechin, I, Zhang, Y, Wang, R, Batchelor, J.D, Mclean, L. | | Deposit date: | 2016-11-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.948 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
