7EH2
 
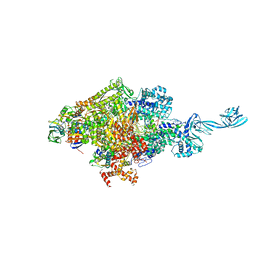 | |
7EH1
 
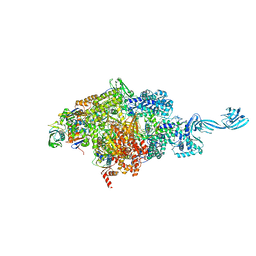 | | Thermus thermophilus transcription initiation complex containing a template-strand purine at position TSS-2, GpG RNA primer, and CMPcPP | | Descriptor: | 1,4-BUTANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (27-MER), ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7CGE
 
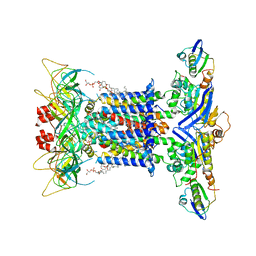 | | The overall structure of nucleotide free MlaFEDB complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CGN
 
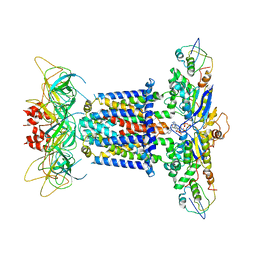 | | The overall structure of the MlaFEDB complex in ATP-bound EQtall conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7VD8
 
 | |
7CH0
 
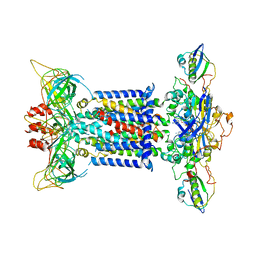 | | The overall structure of the MlaFEDB complex in ATP-bound EQclose conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7C8K
 
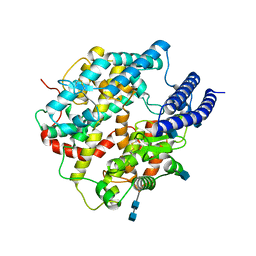 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EB2
 
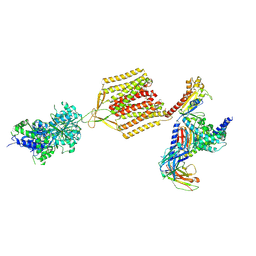 | | Cryo-EM structure of human GABA(B) receptor-Gi protein complex | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Shen, C, Mao, C, Xu, C, Jin, N, Zhang, H, Shen, D, Shen, Q, Wang, X, Hou, T, Rondard, P, Chen, Z, Pin, J, Zhang, Y, Liu, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of GABA B receptor-G i protein coupling.
Nature, 594, 2021
|
|
7DSL
 
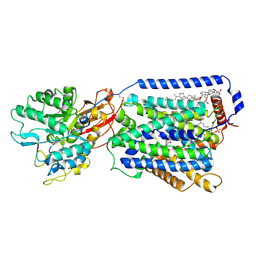 | | Overall structure of the LAT1-4F2hc bound with JX-078 | | Descriptor: | (2~{S})-2-azanyl-7-[(2-phenylphenyl)methoxy]-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSQ
 
 | | Overall structure of the LAT1-4F2hc bound with 3,5-diiodo-L-tyrosine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-DIIODOTYROSINE, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSK
 
 | | Overall structure of the LAT1-4F2hc bound with JX-075 | | Descriptor: | (2~{S})-2-azanyl-7-(naphthalen-1-ylmethoxy)-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
7DSN
 
 | | Overall structure of the LAT1-4F2hc bound with JX-119 | | Descriptor: | (2~{S})-2-azanyl-7-[[2-(1,3-benzoxazol-2-yl)phenyl]methoxy]-3,4-dihydro-1~{H}-naphthalene-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Zhong, X.Y, Zhou, Q. | | Deposit date: | 2020-12-31 | | Release date: | 2021-03-10 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter.
Cell Discov, 7, 2021
|
|
8IXW
 
 | |
7VF9
 
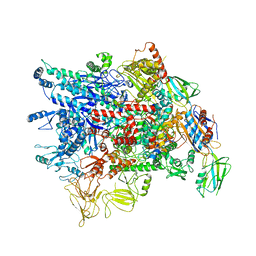 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
8JCB
 
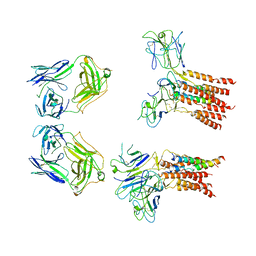 | | Vgamma5 Vdelta1 T cell receptor complex | | Descriptor: | T cell receptor delta variable 1,T cell receptor delta constant, T cell receptor gamma variable 5,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
7EPZ
 
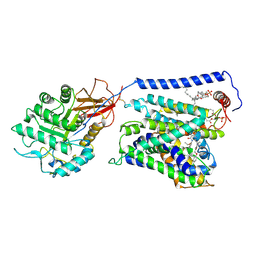 | | Overall structure of Erastin-bound xCT-4F2hc complex | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-[(1S)-1-[4-[2-(4-chloranylphenoxy)ethanoyl]piperazin-1-yl]ethyl]-3-(2-ethoxyphenyl)quinazolin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis.
Cell Res., 32, 2022
|
|
8JC0
 
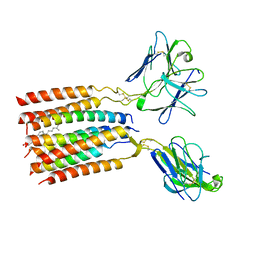 | | V gamma9 V delta2 TCR and CD3 complex in LMNG | | Descriptor: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
7XL4
 
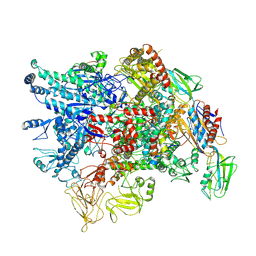 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
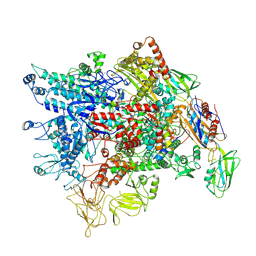 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7WDM
 
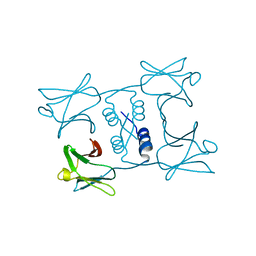 | | Fungal immunomodulatory protein FIP-gmi | | Descriptor: | Immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
7CH1
 
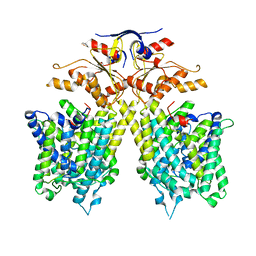 | | The overall structure of SLC26A9 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 26 member 9 | | Authors: | Chi, X.M, Chen, Y, Li, X.R, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the gating mechanism of human SLC26A9 mediated by its C-terminal sequence.
Cell Discov, 6, 2020
|
|
7VDE
 
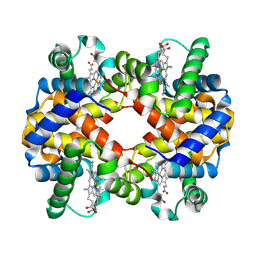 | | 3.6 A structure of the human hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, H.C, Zhang, Y, Sun, F. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI.
Nat Commun, 12, 2021
|
|
7VDA
 
 | |
7VDC
 
 | |
