5LNV
 
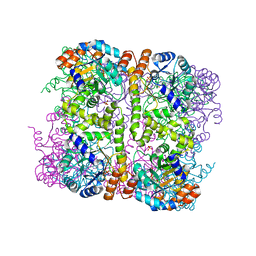 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals | | 分子名称: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | 著者 | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | 登録日 | 2016-08-06 | | 公開日 | 2017-01-18 | | 最終更新日 | 2018-09-19 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5XM7
 
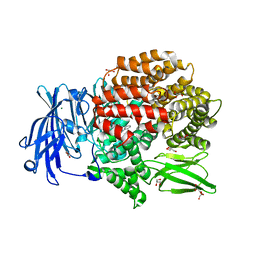 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2S)-4-methyl-N-[(1R)-2-(oxidanylamino)-2-oxidanylidene-1-phenyl-ethyl]-2-[(phenylmethyl)carbamoylamino]pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-05-12 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y1T
 
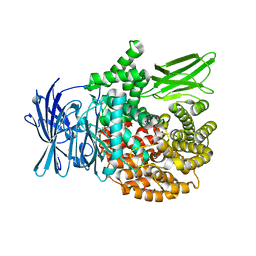 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,3-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2S)-2-[(2,3-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y1K
 
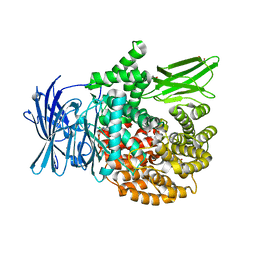 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2S)-2-[(2-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y1Q
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2S)-2-[(3-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, M1 family aminopeptidase, MAGNESIUM ION, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5LNT
 
 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex | | 分子名称: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.1, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | 著者 | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | 登録日 | 2016-08-06 | | 公開日 | 2017-01-18 | | 最終更新日 | 2017-02-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
7V1N
 
 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | 分子名称: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | 著者 | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | 登録日 | 2021-08-05 | | 公開日 | 2022-02-23 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
7V35
 
 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | | 分子名称: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | 登録日 | 2021-08-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
5BTY
 
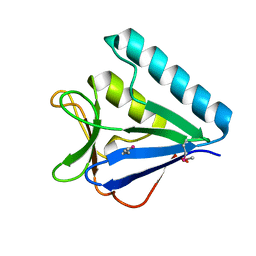 | | Structure of the middle domain of lpg1496 from Legionella pneumophila in P21 space group | | 分子名称: | lpg1496 | | 著者 | Kozlov, G, Zhang, Y, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2015-06-03 | | 公開日 | 2015-08-26 | | 最終更新日 | 2020-01-08 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
3LZD
 
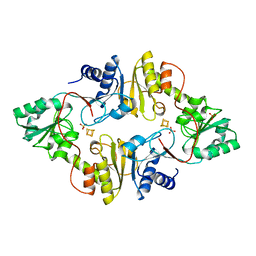 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | 分子名称: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | 著者 | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | 登録日 | 2010-03-01 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
3NQX
 
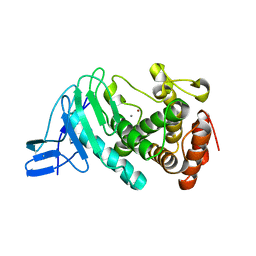 | | Crystal structure of vibriolysin MCP-02 mature enzyme, a zinc metalloprotease from M4 family | | 分子名称: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | 著者 | Gao, X, Wang, J, Wu, J.-W, Zhang, Y.-Z. | | 登録日 | 2010-06-30 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NQZ
 
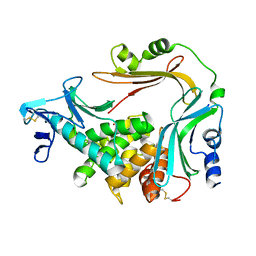 | | Crystal structure of the autoprocessed Vibriolysin MCP-02 with E369A mutation | | 分子名称: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | 著者 | Gao, X, Wang, J, Chen, L, Wu, J.-W, Zhang, Y.-Z. | | 登録日 | 2010-06-30 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JSG
 
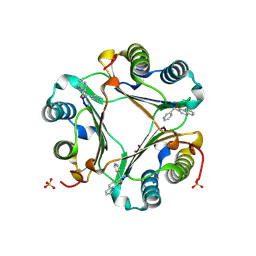 | |
3NQY
 
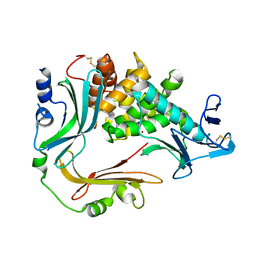 | | Crystal structure of the autoprocessed complex of Vibriolysin MCP-02 with a single point mutation E346A | | 分子名称: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | 著者 | Gao, X, Wang, J, Wu, J.-W, Zhang, Y.-Z. | | 登録日 | 2010-06-30 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JTU
 
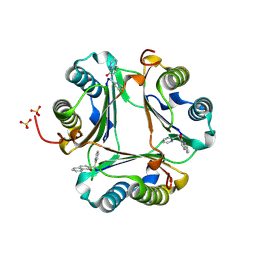 | |
3JCS
 
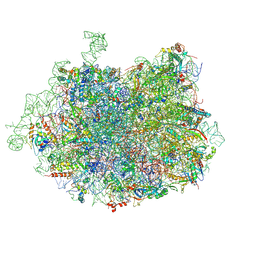 | | 2.8 Angstrom cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania | | 分子名称: | 26S alpha ribosomal RNA, 26S delta ribosomal RNA, 26S epsilon ribosomal RNA, ... | | 著者 | Shalev-Benami, M, Zhang, Y, Matzov, D, Halfon, Y, Zackay, A, Rozenberg, H, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | 登録日 | 2016-01-21 | | 公開日 | 2016-07-20 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | 2.8- angstrom Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania.
Cell Rep, 16, 2016
|
|
3JSF
 
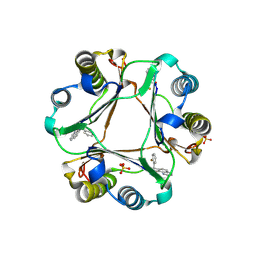 | |
8GTO
 
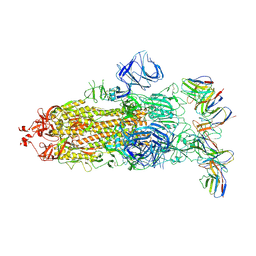 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv282 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GUG
 
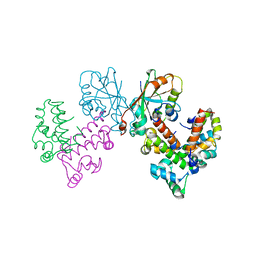 | | Structure of VPA0770 toxin bound to VPA0769 antitoxin in Vibrio parahaemolyticus | | 分子名称: | DUF2384 domain-containing protein, RES domain-containing protein | | 著者 | Song, X.J, Zhang, Y, Xu, Y.Y, Lin, Z. | | 登録日 | 2022-09-12 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural insights of the toxin-antitoxin system VPA0770-VPA0769 in Vibrio parahaemolyticus.
Int.J.Biol.Macromol., 242, 2023
|
|
3L5T
 
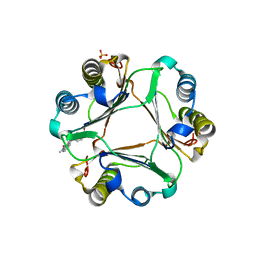 | |
3L5R
 
 | |
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2016-07-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
3L5U
 
 | |
5KPV
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure II) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2016-07-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPW
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | 登録日 | 2016-07-05 | | 公開日 | 2016-09-28 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
